
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,062,034 – 19,062,244 |
| Length | 210 |
| Max. P | 0.998932 |

| Location | 19,062,034 – 19,062,148 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.13 |
| Mean single sequence MFE | -42.64 |
| Consensus MFE | -38.34 |
| Energy contribution | -38.22 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.29 |
| SVM RNA-class probability | 0.998932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
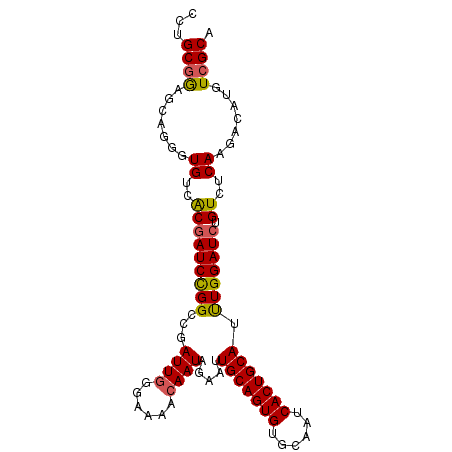
>X_DroMel_CAF1 19062034 114 + 22224390 AACGAAGCGAAAGUGAUGGCCUCACGUUGGGCGCCAUUUGGUGAAUCAGA------UAGUUCCACGUUGGGCGCCACCUAAUGGACCAGAUAAAUGGAUCGAUAGGAGGCCAAGGCGUUA ((((..((....))..(((((((..((((((((((...((..((((....------..))))..))...)))))).((....)).(((......)))..))))..)))))))...)))). ( -42.80) >DroSec_CAF1 13917 114 + 1 AACGAAGCGAAAGUGAUGGCCCCACGUUGGGCGCCAUUUGCUGGUUCAGA------UAGUCCCACGUUGGGCGCCACCUAAUGGACCAGAUAAAUGGAUCGAUAGGAGGCCAAGGCGUUG ((((..((....))..(((((((..(((((...(((((((((((((((..------..(((((.....))).)).......)))))))).))))))).))))).)).)))))...)))). ( -43.80) >DroSim_CAF1 15366 114 + 1 AACGAAGCGAAAGUGAUGGCCCCACGUUGGGCGCCAUUUGCUGGAUCAGA------UAGUCGCACGUUGGGCGCCACCUAAUGGACCAGAUAAAUGGAUCGAUAGGAGGCCAAGGCGUUG ((((..((....))..(((((((..(((((...(((((((((((.((.(.------....)...(((((((.....))))))))))))).))))))).))))).)).)))))...)))). ( -41.10) >DroEre_CAF1 14248 120 + 1 AACGAAGCGAAAGUGAUGGCCCCACGUUGGGCGCCAUUUGGUGGACCAGAGCAAAGUAGUCCCACGUUGGGCGCCACUUAAUGGAGCAGAUAAAUGGAUCGAUAGGAGGCCAAGGCGUUA ((((..((....))..(((((((.(((((((((((...((..((((............))))..))...))))))...))))).....(((......)))....)).)))))...)))). ( -39.30) >DroYak_CAF1 15414 114 + 1 AACGAAGCGAAAGUGAUGGCCCCACGUUGGGCGCCACUUGUUGGACCUGA------UAGUCCCACGUUGGGCGCCACCUAAUGGACCAGAUAAAUGGAUCGAUAGGAGGCCAAGGCGUUA ((((..((....))..(((((........((((((...(((.((((....------..)))).)))...)))))).((((.(((.(((......))).))).)))).)))))...)))). ( -46.20) >consensus AACGAAGCGAAAGUGAUGGCCCCACGUUGGGCGCCAUUUGCUGGAUCAGA______UAGUCCCACGUUGGGCGCCACCUAAUGGACCAGAUAAAUGGAUCGAUAGGAGGCCAAGGCGUUA ((((..((....))..(((((........((((((...((..((((............))))..))...)))))).((((.(((.(((......))).))).)))).)))))...)))). (-38.34 = -38.22 + -0.12)


| Location | 19,062,148 – 19,062,244 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 92.81 |
| Mean single sequence MFE | -30.54 |
| Consensus MFE | -24.66 |
| Energy contribution | -24.62 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19062148 96 + 22224390 CCUGCGGAGCAUGGUGUCACGAUCCGGUCGAUUGGGAAAACAAUAGAAUUGCAGUGUGCAAUCACUGCAUUUGGAUCUGUCUCAAGACAUGUCGCA ...(((..(((((.((..(((((((((((.((((......)))).))..(((((((......))))))).))))))).))..))...)))))))). ( -32.70) >DroSec_CAF1 14031 96 + 1 CCUGCGGAGCAGGGUGUCACGAUCCGGCCGAUUGGGAAAACAAUAGAAUUGCAGUGUGCAAUCACUGCAUUUGGAUCUGUCUCAAGACAUGUCGCA (((((...)))))(((.((.(((((((...((((......)))).....(((((((......))))))).)))))))((((....)))))).))). ( -33.90) >DroSim_CAF1 15480 96 + 1 CCUGCGGAGCAGGGUGUCACGAUCCGGCCGAUUGGGAAAACAAUAGAAUUGCAGUGUGCAAUCACUGCAUUUGGAUCUGUCUCAAGACAUGUCGCA (((((...)))))(((.((.(((((((...((((......)))).....(((((((......))))))).)))))))((((....)))))).))). ( -33.90) >DroEre_CAF1 14368 96 + 1 CCUGCGGAGCAGAGUGUCGCGAUCUGGCCGAUUCGGAAAACAAUAGAAUUGCAGUGCGCAAUCACUGCAUUUGGAUAUGUCUCAACAGAUGUCGCA .((((...))))......((((((((.(((...)))...(((((..((.(((((((......))))))).))..)).))).....))))..)))). ( -24.90) >DroYak_CAF1 15528 96 + 1 CCUGCCAAGCAGUGUGUCACGAUCCGGCCGAUUGGAAAAACAAUAGAAUUGCAGUGUGCAAUCACUGCAUCUGGAUCUGUUCCAAGAGAUGUCGCA .((((...))))((((.((.(((((((...((((......)))).....(((((((......))))))).)))))))...((.....)))).)))) ( -27.30) >consensus CCUGCGGAGCAGGGUGUCACGAUCCGGCCGAUUGGGAAAACAAUAGAAUUGCAGUGUGCAAUCACUGCAUUUGGAUCUGUCUCAAGACAUGUCGCA ...((((.......((..(((((((((...((((......)))).....(((((((......))))))).))))))).))..)).......)))). (-24.66 = -24.62 + -0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:18 2006