
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,061,504 – 19,061,714 |
| Length | 210 |
| Max. P | 0.941393 |

| Location | 19,061,504 – 19,061,599 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 82.05 |
| Mean single sequence MFE | -21.27 |
| Consensus MFE | -14.51 |
| Energy contribution | -14.82 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
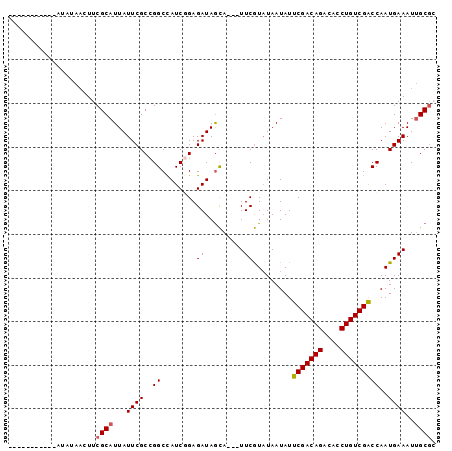
>X_DroMel_CAF1 19061504 95 + 22224390 ACAAUACAUAUAUAUAACUUCGCAUUAUUCGCCGGCCAUCUGAGAUAGCA---UUCGUCUAAUAUUCGACAGACACCUGUCGACCAAUGAAAUUGCAG .....................(((...((((..((.......((((....---...)))).....(((((((....)))))))))..))))..))).. ( -17.30) >DroSec_CAF1 13398 84 + 1 -----------AUAUAACUUCGCAUUAUUCGCCGGCCAUCGGAGAUAGCA---UUCGUAUAAUAUUCGACAGACACCUGUCGGCCAAUGAAAUUGCGC -----------.........((((...((((..((((..((((.......---))))..........(((((....)))))))))..))))..)))). ( -23.20) >DroSim_CAF1 14844 87 + 1 -----------AUAUAACUUCGCAUUAUUCGCCGGCCAUCGGAGAUAAUAUAUUUCAUAUAAUAUUCGACAGACACCUGUCGACCAAUGAAAUUGCGC -----------.........(((((((((..(((.....))).))))))..(((((((.......(((((((....)))))))...))))))).))). ( -20.40) >DroEre_CAF1 13740 77 + 1 ------------------UUCGCAUUAUUCGCUGGCUAUCUGAGAUAGCA---UUCGUAUAAUAUUCGACAGACACCUGUCGACCAAUGAAAUCGCGG ------------------..((((((((.((...((((((...)))))).---..)).)))))..(((((((....)))))))...........))). ( -24.20) >consensus ___________AUAUAACUUCGCAUUAUUCGCCGGCCAUCGGAGAUAGCA___UUCGUAUAAUAUUCGACAGACACCUGUCGACCAAUGAAAUUGCGC ....................((((...((((..((........((.........)).........(((((((....)))))))))..))))..)))). (-14.51 = -14.82 + 0.31)


| Location | 19,061,504 – 19,061,599 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 82.05 |
| Mean single sequence MFE | -23.43 |
| Consensus MFE | -19.96 |
| Energy contribution | -19.27 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.810489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
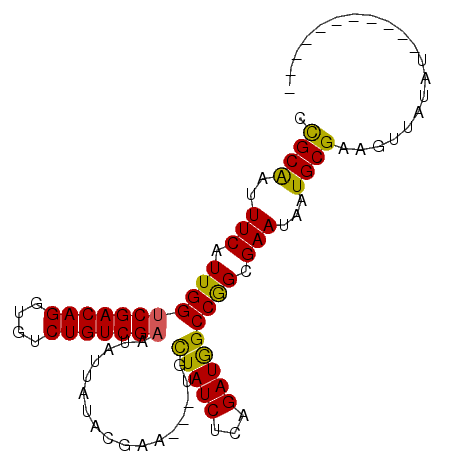
>X_DroMel_CAF1 19061504 95 - 22224390 CUGCAAUUUCAUUGGUCGACAGGUGUCUGUCGAAUAUUAGACGAA---UGCUAUCUCAGAUGGCCGGCGAAUAAUGCGAAGUUAUAUAUAUGUAUUGU ....((((((.....(((((((....))))))).(((((..((..---.((((((...))))))...))..))))).))))))............... ( -23.30) >DroSec_CAF1 13398 84 - 1 GCGCAAUUUCAUUGGCCGACAGGUGUCUGUCGAAUAUUAUACGAA---UGCUAUCUCCGAUGGCCGGCGAAUAAUGCGAAGUUAUAU----------- .((((..(((.(((((((((((....)))))..........((..---.........))..)))))).)))...)))).........----------- ( -22.70) >DroSim_CAF1 14844 87 - 1 GCGCAAUUUCAUUGGUCGACAGGUGUCUGUCGAAUAUUAUAUGAAAUAUAUUAUCUCCGAUGGCCGGCGAAUAAUGCGAAGUUAUAU----------- .(((((((((((...(((((((....))))))).......)))))))......((.(((.....))).))....)))).........----------- ( -23.90) >DroEre_CAF1 13740 77 - 1 CCGCGAUUUCAUUGGUCGACAGGUGUCUGUCGAAUAUUAUACGAA---UGCUAUCUCAGAUAGCCAGCGAAUAAUGCGAA------------------ .(((...........(((((((....)))))))..(((((.((..---.((((((...))))))...)).))))))))..------------------ ( -23.80) >consensus CCGCAAUUUCAUUGGUCGACAGGUGUCUGUCGAAUAUUAUACGAA___UGCUAUCUCAGAUGGCCGGCGAAUAAUGCGAAGUUAUAU___________ .((((..(((.(((((((((((....))))))).................(((((...))))))))).)))...)))).................... (-19.96 = -19.27 + -0.69)


| Location | 19,061,599 – 19,061,714 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 93.13 |
| Mean single sequence MFE | -28.08 |
| Consensus MFE | -23.58 |
| Energy contribution | -23.66 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.860294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19061599 115 + 22224390 ACAGACUAAAGGCGAGAUAAGUCGAUCAUCGAUAAAAGUCACUUCGAAAGUAUUCGUAAUUAUAAUUUGAGUUGUAAGUGGCCCACGGUGGAAGACCAUAAAGGCCUGAGGUCGA ...((((..((((.......(((..((((((......(((((..((((....))))...(((((((....))))))))))))...))))))..))).......))))..)))).. ( -30.14) >DroSec_CAF1 13482 115 + 1 ACAGACUAAAGGCGAGAUAAAUCGAUCAUCGAUAAAAGUCACUUCGAAAGUAUGCGUAAAUAUAAUUUGAGUUGUAAGUGGCCCACGGUGGAAGACCAUAAAGGCCUGAGGUCGA ...((((..((((..(((..((((.....))))....))).((((.(..((..((.((..((((((....))))))..))))..))..).)))).........))))..)))).. ( -27.00) >DroSim_CAF1 14931 115 + 1 ACAGACUGAAGGCGAGAUAAAUCGAUCAUCGAUAAAAGUCACUUCGAAAGUAUGCGUAAAUAUAAUUUGAGUUGUAAGUGGCCCACGGUGGAAGACUAUAAAGGCCUGAGGUCGA ...((((..((((..(((..((((.....))))....))).((((.(..((..((.((..((((((....))))))..))))..))..).)))).........))))..)))).. ( -28.00) >DroEre_CAF1 13817 115 + 1 ACAGACUGAAGGCGAGAUAAGUCGAUCAUCGAUAAAAGUCACUUUGAAAGUAUUCGUAAUUAUAAUUUGAGUUGUAAAUGCCACACGGUGGAAGACCAUAAAAGCCUAAGGUCGA ...((((..((((.......(((..((((((.......((.....))..(((((..(((((........)))))..)))))....))))))..))).......))))..)))).. ( -28.04) >DroYak_CAF1 14945 115 + 1 ACAGACUGAAGGCGAGAUAAGGCGAUCAUCGAUAAAAGUCACUUCGAAAGUAUUCGUAAUUAUAAUUUGAGUUAUAAGAGGCGCAUGAUGGAAGACCAUAAAGGCCUGAGGUCGU ...((((..((((........(((.((.((((...........))))............(((((((....)))))))))..)))...((((....))))....))))..)))).. ( -27.20) >consensus ACAGACUGAAGGCGAGAUAAGUCGAUCAUCGAUAAAAGUCACUUCGAAAGUAUUCGUAAUUAUAAUUUGAGUUGUAAGUGGCCCACGGUGGAAGACCAUAAAGGCCUGAGGUCGA ...((((..((((..(((..((((.....))))....))).((((.(..((....((..(((((((....)))))))...))..))..).)))).........))))..)))).. (-23.58 = -23.66 + 0.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:16 2006