
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,057,891 – 19,057,982 |
| Length | 91 |
| Max. P | 0.993069 |

| Location | 19,057,891 – 19,057,982 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 88.90 |
| Mean single sequence MFE | -19.71 |
| Consensus MFE | -13.57 |
| Energy contribution | -14.37 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
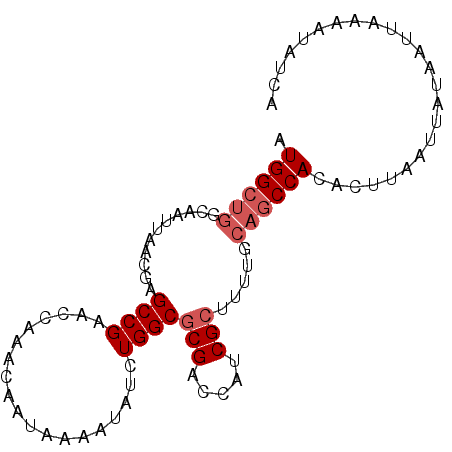
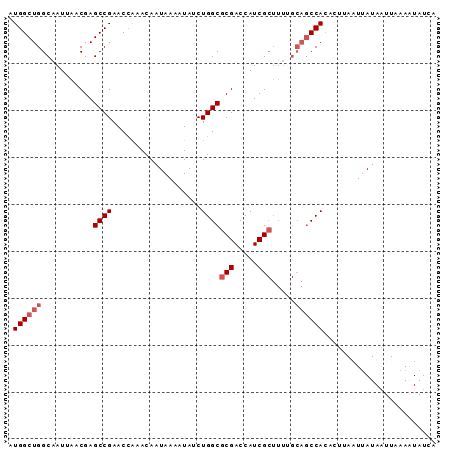
>X_DroMel_CAF1 19057891 91 + 22224390 AUGGCUGGCAAUUAACGAGCCGAACCAAACAAUAAAAUAUCUGGCGCGACCAUCGCUUUUGCAGCCACACUUAAUUAUAAUUUAAGUAUCA .((((((.(((.......((((...................))))(((.....)))..))))))))).((((((.......)))))).... ( -21.81) >DroSec_CAF1 9713 91 + 1 AUGGCUGGCAAUUAACGAGCCGAACCAAACAAUAAAAUAACUGGCGCGACCAUCGCUUUUGCAGCCACACUUAAUUAUAAUUAAAAUAUCA .((((((.(((.......((((...................))))(((.....)))..)))))))))........................ ( -18.71) >DroSim_CAF1 11257 91 + 1 AUGGCUGGCAAUUAACGAGCCGAACCGAACAAUAAAAUAACUGGCGCGACCAUCGCUUUGGCAGCCACACUUAAUUAUAAUUAAAAUAUCA .((((((.(((.......((((...................))))(((.....))).))).))))))........................ ( -21.21) >DroEre_CAF1 10351 81 + 1 AUGGCUGGCAAUUAACGAGCCGAACCAAACAAUAAAAUAUCUGGCGCGACCAUCG---------CCAGAUUAA-UUAUACUUCAAAUAUUA .(((((.(.......).))))).........((((...((((((((.......))---------))))))...-))))............. ( -19.50) >DroYak_CAF1 11327 91 + 1 AUGGCUGGCAAUUAACGAGCCGAACGAAACAAUAAAAUAUCUGGCGCGACCAUCGCUUUAGCAGCCACACUAAGUCAUAAUUAAAAUAUUA .((((((.(.........((((...................))))(((.....)))....)))))))........................ ( -17.31) >consensus AUGGCUGGCAAUUAACGAGCCGAACCAAACAAUAAAAUAUCUGGCGCGACCAUCGCUUUUGCAGCCACACUUAAUUAUAAUUAAAAUAUCA .((((((...........((((...................))))(((.....))).....))))))........................ (-13.57 = -14.37 + 0.80)

| Location | 19,057,891 – 19,057,982 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 88.90 |
| Mean single sequence MFE | -26.26 |
| Consensus MFE | -20.86 |
| Energy contribution | -22.86 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.993069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
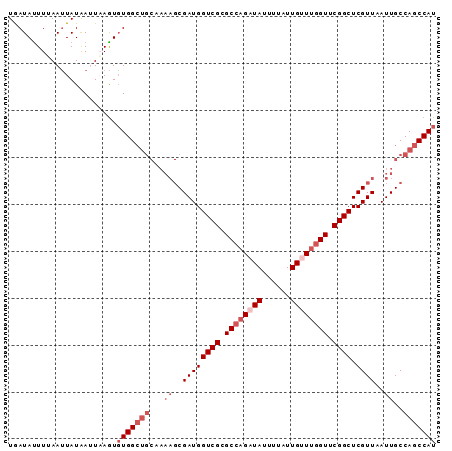
>X_DroMel_CAF1 19057891 91 - 22224390 UGAUACUUAAAUUAUAAUUAAGUGUGGCUGCAAAAGCGAUGGUCGCGCCAGAUAUUUUAUUGUUUGGUUCGGCUCGUUAAUUGCCAGCCAU ..((((((((.......))))))))((((((((.(((((..((((.((((((((......)))))))).)))))))))..))).))))).. ( -33.90) >DroSec_CAF1 9713 91 - 1 UGAUAUUUUAAUUAUAAUUAAGUGUGGCUGCAAAAGCGAUGGUCGCGCCAGUUAUUUUAUUGUUUGGUUCGGCUCGUUAAUUGCCAGCCAU .......................((((((((((.(((((..((((.(((((.((......)).))))).)))))))))..))).))))))) ( -26.20) >DroSim_CAF1 11257 91 - 1 UGAUAUUUUAAUUAUAAUUAAGUGUGGCUGCCAAAGCGAUGGUCGCGCCAGUUAUUUUAUUGUUCGGUUCGGCUCGUUAAUUGCCAGCCAU .......................(((((((.((((((((..((((.(((...((......))...))).)))))))))..))).))))))) ( -23.30) >DroEre_CAF1 10351 81 - 1 UAAUAUUUGAAGUAUAA-UUAAUCUGG---------CGAUGGUCGCGCCAGAUAUUUUAUUGUUUGGUUCGGCUCGUUAAUUGCCAGCCAU .................-.....((((---------(((((((((.((((((((......)))))))).))))).....)))))))).... ( -25.50) >DroYak_CAF1 11327 91 - 1 UAAUAUUUUAAUUAUGACUUAGUGUGGCUGCUAAAGCGAUGGUCGCGCCAGAUAUUUUAUUGUUUCGUUCGGCUCGUUAAUUGCCAGCCAU .......................(((((((.((((((((..((((.((.(((((......))))).)).)))))))))..))).))))))) ( -22.40) >consensus UGAUAUUUUAAUUAUAAUUAAGUGUGGCUGCAAAAGCGAUGGUCGCGCCAGAUAUUUUAUUGUUUGGUUCGGCUCGUUAAUUGCCAGCCAU .......................(((((((...((..((((((((.((((((((......)))))))).)))).))))..))..))))))) (-20.86 = -22.86 + 2.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:09 2006