
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,052,466 – 19,052,620 |
| Length | 154 |
| Max. P | 0.989530 |

| Location | 19,052,466 – 19,052,586 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.19 |
| Mean single sequence MFE | -22.64 |
| Consensus MFE | -20.20 |
| Energy contribution | -20.40 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19052466 120 + 22224390 UCAUGCUCAUCCCCAUUCAAAUACCCAACCGCCUUCCUUCCAAAUCGCAAUCGUAAAACCCUGGAUACGUGCAUAGUGCCAUUUGGCACACAAGCUGUUGCCAUUUGGCCCUAAACGAUC ...........................................((((((..((((...(....).))))))).(((.((((..(((((.((.....)))))))..)))).)))...))). ( -22.90) >DroSec_CAF1 4320 119 + 1 UCAUGCUCAUCCCCAUCCAAAUACCCAACCGCCUUCC-ACCAAAUCGCAAUCAUAAAACCCUGGAUACGUGCAUAGUGCCAUUUGGCACACAAGCUGUUGCCAUUUGGCCCUAAACGAUC ..............(((((..................-.......................))))).(((...(((.((((..(((((.((.....)))))))..)))).))).)))... ( -22.58) >DroSim_CAF1 5679 119 + 1 UCAUGCUCAUCCCCAUCCAAAUACCCAACCGCCUUCC-ACCAAAUCGCAAUCAUAAAACCCUGGAUACGUGCAUAGUGCCAUUUGGCACACAAGCUGUUGCCAUUUGGCCCUAAACGAUC ..............(((((..................-.......................))))).(((...(((.((((..(((((.((.....)))))))..)))).))).)))... ( -22.58) >DroEre_CAF1 5316 109 + 1 NNNNNNNNNCCCCCAUCCCCAUCCCCA-----------UCCAAAUCGCAAUCAUAAAACCCUGGAUACGUGCAUAGUGCCAUUUGGCACACAAGCUGUUGCCAUUUGGCCCUAAACGAUC ..........................(-----------((((...................))))).(((...(((.((((..(((((.((.....)))))))..)))).))).)))... ( -23.41) >DroYak_CAF1 5482 105 + 1 --------ACCCC-CUCUCCAAACCCA-----CUCCU-UUGAAAUCGCAAUCAUAAAAGCCUGGAUACGUGCAUAGUGCCAUUUGGCACACAAGCUCUUGCCAUUUGGCCCUAAACGAUC --------.....-...((((......-----.....-(((......)))...........))))..(((...(((.((((..(((((..........)))))..)))).))).)))... ( -21.75) >consensus UCAUGCUCAUCCCCAUCCAAAUACCCAACCGCCUUCC_UCCAAAUCGCAAUCAUAAAACCCUGGAUACGUGCAUAGUGCCAUUUGGCACACAAGCUGUUGCCAUUUGGCCCUAAACGAUC ...............................................................(((.(((...(((.((((..(((((.((.....)))))))..)))).))).)))))) (-20.20 = -20.40 + 0.20)



| Location | 19,052,506 – 19,052,620 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.82 |
| Mean single sequence MFE | -33.78 |
| Consensus MFE | -20.43 |
| Energy contribution | -22.55 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.984836 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19052506 114 + 22224390 CAAAUCGCAAUCGUAAAACCCUGGAUACGUGCAUAGUGCCAUUUGGCACACAAGCUGUUGCCAUUUGGCCCUAAACGAUCAUAGGCAAAUGGAUAGUUAACUGAGUUCGCGA------AC ....((((...........(((((((.(((...(((.((((..(((((.((.....)))))))..)))).))).)))))).)))).....((((((....))..))))))))------.. ( -33.30) >DroPse_CAF1 5873 87 + 1 --G----------CUAAUCUCUGGGUACGUGC-UAGUCCGAGUAG----------UACCGCCAUUUGGCCCUAAACGGUCAUAGGCAAACAAACAGCAGAUUGAGAUUGA---------- --.----------.((((((((((((((.(((-(......)))))----------))))(((...(((((......)))))..))).......))).)))))).......---------- ( -29.00) >DroSec_CAF1 4359 114 + 1 CAAAUCGCAAUCAUAAAACCCUGGAUACGUGCAUAGUGCCAUUUGGCACACAAGCUGUUGCCAUUUGGCCCUAAACGAUCGUAGGCAAUUGGAUAGUUAACUGAGUUUGCGA------AC ....((((((.(.......(((((((.(((...(((.((((..(((((.((.....)))))))..)))).))).)))))).))))........(((....))).).))))))------.. ( -34.70) >DroSim_CAF1 5718 114 + 1 CAAAUCGCAAUCAUAAAACCCUGGAUACGUGCAUAGUGCCAUUUGGCACACAAGCUGUUGCCAUUUGGCCCUAAACGAUCGUAGGCAAAUGGAUAGUUAACUGAGUUUGCGA------AC ....((((((.(.......(((((((.(((...(((.((((..(((((.((.....)))))))..)))).))).)))))).))))........(((....))).).))))))------.. ( -34.70) >DroEre_CAF1 5345 108 + 1 CAAAUCGCAAUCAUAAAACCCUGGAUACGUGCAUAGUGCCAUUUGGCACACAAGCUGUUGCCAUUUGGCCCUAAACGAUCAU------------AGUUAACUGGGUUUGCGAACCCGAAC ....((((((.......((((..(((.(((...(((.((((..(((((.((.....)))))))..)))).))).))))))..------------((....))))))))))))........ ( -33.31) >DroYak_CAF1 5507 114 + 1 GAAAUCGCAAUCAUAAAAGCCUGGAUACGUGCAUAGUGCCAUUUGGCACACAAGCUCUUGCCAUUUGGCCCUAAACGAUCAUAGGCAAAUGAGUAGUUAACUGAGUUUGCGA------AC ....((((((.(......((((((((.(((...(((.((((..(((((..........)))))..)))).))).)))))).)))))...(.(((.....))).)).))))))------.. ( -37.70) >consensus CAAAUCGCAAUCAUAAAACCCUGGAUACGUGCAUAGUGCCAUUUGGCACACAAGCUGUUGCCAUUUGGCCCUAAACGAUCAUAGGCAAAUGGAUAGUUAACUGAGUUUGCGA______AC ....((((((.(.......(((((((.(((...(((.((((..(((((..........)))))..)))).))).)))))).))))........(((....))).).))))))........ (-20.43 = -22.55 + 2.12)

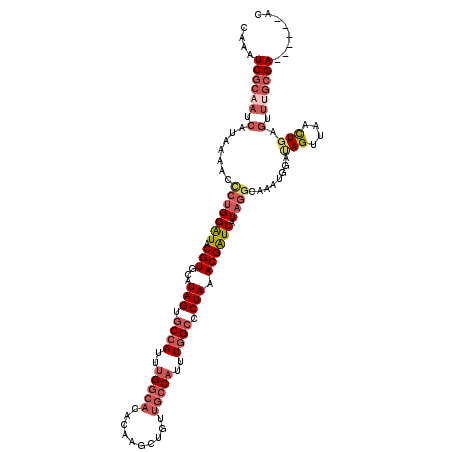

| Location | 19,052,506 – 19,052,620 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.82 |
| Mean single sequence MFE | -30.58 |
| Consensus MFE | -19.78 |
| Energy contribution | -21.06 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560734 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19052506 114 - 22224390 GU------UCGCGAACUCAGUUAACUAUCCAUUUGCCUAUGAUCGUUUAGGGCCAAAUGGCAACAGCUUGUGUGCCAAAUGGCACUAUGCACGUAUCCAGGGUUUUACGAUUGCGAUUUG ..------((((((..............((((((((((.(((....)))))).))))))).....((....(((((....)))))...)).((((.(....)...)))).)))))).... ( -31.30) >DroPse_CAF1 5873 87 - 1 ----------UCAAUCUCAAUCUGCUGUUUGUUUGCCUAUGACCGUUUAGGGCCAAAUGGCGGUA----------CUACUCGGACUA-GCACGUACCCAGAGAUUAG----------C-- ----------..((((((.....((((((((...((((...........))))))))))))((((----------(...........-....)))))..))))))..----------.-- ( -22.96) >DroSec_CAF1 4359 114 - 1 GU------UCGCAAACUCAGUUAACUAUCCAAUUGCCUACGAUCGUUUAGGGCCAAAUGGCAACAGCUUGUGUGCCAAAUGGCACUAUGCACGUAUCCAGGGUUUUAUGAUUGCGAUUUG ..------((((((..(((...((((......(((..((((...((.(((.((((..(((((((.....)).)))))..)))).))).)).))))..)))))))...))))))))).... ( -30.90) >DroSim_CAF1 5718 114 - 1 GU------UCGCAAACUCAGUUAACUAUCCAUUUGCCUACGAUCGUUUAGGGCCAAAUGGCAACAGCUUGUGUGCCAAAUGGCACUAUGCACGUAUCCAGGGUUUUAUGAUUGCGAUUUG ..------((((((..(((...((((..((((((((((((....))...))).))))))).....((....(((((....)))))...))..........))))...))))))))).... ( -33.30) >DroEre_CAF1 5345 108 - 1 GUUCGGGUUCGCAAACCCAGUUAACU------------AUGAUCGUUUAGGGCCAAAUGGCAACAGCUUGUGUGCCAAAUGGCACUAUGCACGUAUCCAGGGUUUUAUGAUUGCGAUUUG ....(((((....)))))........------------..((((((.(((((((...(((..((.((....(((((....)))))...))..))..))).))))))).....)))))).. ( -31.50) >DroYak_CAF1 5507 114 - 1 GU------UCGCAAACUCAGUUAACUACUCAUUUGCCUAUGAUCGUUUAGGGCCAAAUGGCAAGAGCUUGUGUGCCAAAUGGCACUAUGCACGUAUCCAGGCUUUUAUGAUUGCGAUUUC ..------((((((....((....))..((((..((((..((((((.....(((....)))....((....(((((....)))))...))))).))).))))....)))))))))).... ( -33.50) >consensus GU______UCGCAAACUCAGUUAACUAUCCAUUUGCCUAUGAUCGUUUAGGGCCAAAUGGCAACAGCUUGUGUGCCAAAUGGCACUAUGCACGUAUCCAGGGUUUUAUGAUUGCGAUUUG ........((((((((((.(((.............((((........))))(((....)))))).((....(((((....)))))...)).........))))).......))))).... (-19.78 = -21.06 + 1.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:02 2006