| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,044,718 – 19,044,909 |
| Length | 191 |
| Max. P | 0.955497 |

| Location | 19,044,718 – 19,044,829 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 81.10 |
| Mean single sequence MFE | -35.40 |
| Consensus MFE | -20.86 |
| Energy contribution | -23.67 |
| Covariance contribution | 2.81 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19044718 111 + 22224390 AGAUGACA-GAUGAUUAGCCCCAA-CUUGUUACGGAUG-ACUGAAUCCCAGGUAACUUACCUGCAAAGAAUCCGUAGAAGACAAUAGCUACCACCUGUGUGGUGGUUGUGACCA ........-...............-(((.((((((((.-.((......((((((...))))))...)).)))))))))))...(((((((((((....)))))))))))..... ( -31.10) >DroSec_CAF1 1848 111 + 1 GGAUGACA-GAUGAUUAGGCAAAA-CUGGUUACGGAUG-UCUGAAUCCCAGGUAACUUACCUGCAAAGAAUCCGUAGAAGACAAUAGCCACCACCUGUGUGGUGGUGGUGACCA ((.((...-...((((((......-))))))((((((.-(((......((((((...))))))...)))))))))......))...(((((((((.....)))))))))..)). ( -38.00) >DroSim_CAF1 1935 111 + 1 GGAUGACA-GAUGAUUAGGCAAAA-CUGGUUACGGAUG-ACUGAAUCCCAGGUAACUUACCUGCAAAGAAUCCGUAGAAGACAAUGGCCACCACCUGUGUGGUGGUGGUGACCA ((....((-..(((((((......-)))))(((((((.-.((......((((((...))))))...)).))))))).....)).))(((((((((.....)))))))))..)). ( -37.30) >DroEre_CAF1 5575 108 + 1 AGACGACA-GA---UUAGGCAAAA-CUGUUUGCGGAUG-ACUGAGGCCCAGGUAACUUACCUGCAAAGCAUCCGUAGAAGACAAUGGCCACCACCUGCGUGGUGGUGGUGACCA ........-..---...(((....-((.((((((((((-.((...((..(((((...)))))))..))))))))))))))......(((((((((.....)))))))))).)). ( -41.10) >DroYak_CAF1 2001 110 + 1 AGAUGACA-GA---UUAGGCAAAAGCUGGUUACGCAUAAACUGAAUCCCAGGCAACUUACCUGCAAAGCAGCCGUAGAAGACAAUGGCCACCACCUGUGUGGUGGUGGUGACCA ........-..---...(((....)))((((((((.....(((.....)))(((.......)))...)).(((((........)))))(((((((.....))))))))))))). ( -35.30) >DroMoj_CAF1 5147 96 + 1 GGAUGCUGGGAGCAGCAGGCACAG-CUG---------------G--CACAAACGACUUACCCGCAAAGCAGCCGUAGAAGACAAUGGCCACGGUCUGAGUGGUGGUGGUGACCA ...(((((....)))))(((((.(-(((---------------.--(((..(.((((.....((...)).(((((........)))))...)))))..))).)))).))).)). ( -29.60) >consensus AGAUGACA_GAUGAUUAGGCAAAA_CUGGUUACGGAUG_ACUGAAUCCCAGGUAACUUACCUGCAAAGAAUCCGUAGAAGACAAUGGCCACCACCUGUGUGGUGGUGGUGACCA .................((..........((((((((...((......((((((...))))))...)).)))))))).........(((((((((.....)))))))))..)). (-20.86 = -23.67 + 2.81)



| Location | 19,044,718 – 19,044,829 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 81.10 |
| Mean single sequence MFE | -34.48 |
| Consensus MFE | -22.23 |
| Energy contribution | -24.53 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.828662 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19044718 111 - 22224390 UGGUCACAACCACCACACAGGUGGUAGCUAUUGUCUUCUACGGAUUCUUUGCAGGUAAGUUACCUGGGAUUCAGU-CAUCCGUAACAAG-UUGGGGCUAAUCAUC-UGUCAUCU ((((....))))....(((((((((((((.....(((.(((((((.((((.((((((...)))))).))...)).-.)))))))..)))-....)))).))))))-)))..... ( -34.20) >DroSec_CAF1 1848 111 - 1 UGGUCACCACCACCACACAGGUGGUGGCUAUUGUCUUCUACGGAUUCUUUGCAGGUAAGUUACCUGGGAUUCAGA-CAUCCGUAACCAG-UUUUGCCUAAUCAUC-UGUCAUCC (((((((((((........)))))))))))........((((((((((((.((((((...)))))).))...)))-.)))))))..(((-...((......)).)-))...... ( -36.20) >DroSim_CAF1 1935 111 - 1 UGGUCACCACCACCACACAGGUGGUGGCCAUUGUCUUCUACGGAUUCUUUGCAGGUAAGUUACCUGGGAUUCAGU-CAUCCGUAACCAG-UUUUGCCUAAUCAUC-UGUCAUCC (((((((((((........)))))))))))........(((((((.((((.((((((...)))))).))...)).-.)))))))..(((-...((......)).)-))...... ( -36.50) >DroEre_CAF1 5575 108 - 1 UGGUCACCACCACCACGCAGGUGGUGGCCAUUGUCUUCUACGGAUGCUUUGCAGGUAAGUUACCUGGGCCUCAGU-CAUCCGCAAACAG-UUUUGCCUAA---UC-UGUCGUCU (((((((((((........)))))))))))..((..((....)).))....((((((...))))))(((..(((.-.....(((((...-.)))))....---.)-))..))). ( -37.90) >DroYak_CAF1 2001 110 - 1 UGGUCACCACCACCACACAGGUGGUGGCCAUUGUCUUCUACGGCUGCUUUGCAGGUAAGUUGCCUGGGAUUCAGUUUAUGCGUAACCAGCUUUUGCCUAA---UC-UGUCAUCU (((((((((((........)))))))))))........((((((((..((.(((((.....))))).))..)))).....))))....((....))....---..-........ ( -36.30) >DroMoj_CAF1 5147 96 - 1 UGGUCACCACCACCACUCAGACCGUGGCCAUUGUCUUCUACGGCUGCUUUGCGGGUAAGUCGUUUGUG--C---------------CAG-CUGUGCCUGCUGCUCCCAGCAUCC (((((((...(........)...)))))))........(((((((((...((((.....))))....)--.---------------)))-)))))..(((((....)))))... ( -25.80) >consensus UGGUCACCACCACCACACAGGUGGUGGCCAUUGUCUUCUACGGAUGCUUUGCAGGUAAGUUACCUGGGAUUCAGU_CAUCCGUAACCAG_UUUUGCCUAAUCAUC_UGUCAUCC (((((((((((........)))))))))))........(((((((.((((.(((((.....))))).))...))...))))))).............................. (-22.23 = -24.53 + 2.31)


| Location | 19,044,789 – 19,044,909 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.17 |
| Mean single sequence MFE | -42.85 |
| Consensus MFE | -20.03 |
| Energy contribution | -20.07 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.822189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
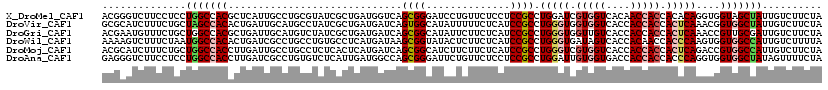
>X_DroMel_CAF1 19044789 120 + 22224390 UAGAAGACAAUAGCUACCACCUGUGUGGUGGUUGUGACCACGAUCCAGGCGGAGGAGAACAGGAUCCCGCUGACCAUCAGCGAUACGCAGGCAAUGAGCGUGGCCAGGAGGAAGACCCGU .........(((((((((((....)))))))))))........(((.(((...(((........)))(((((.....))))).(((((.........)))))))).)))((.....)).. ( -38.60) >DroVir_CAF1 5488 120 + 1 UAGAAGACAAUAGCCACCGUUUGAGUGGUGGUGGUGACCACCACCCAGGCGGAUGAGAAAAAUAUGCCACUGAUCAUCAGCGAUAGGCAUGCAAUCAGUGUGGCUAGCAGAAAGAUGCGC ..........(((((((((((((.((((((((....)))))))).)))))))...............(((((((...((((.....)).))..)))))))))))))(((......))).. ( -46.40) >DroGri_CAF1 6274 120 + 1 UAGAAGACAAUCGCAACGGUUUGAGUGGUGGUGGUGACAACCACCCAGGCGGAUGAGAAGAAUAUGCCGCUGAUCAUCAGCGAUAGACAUGCAAUCAGCGUGGCCAGCAGAAACAUUCGU ..........((.((.(.(((((...(((((((....).))))))))))).).)).)).((((.((((((((.....))))).....(((((.....)))))....))).....)))).. ( -37.00) >DroWil_CAF1 1489 120 + 1 UAAAAGACAAUGGCCACCACUUGGGUGGUUGUGGUGACUAUCACCCAGGCGGAUGAGAAGAGUAUACCGCUUAUCAUGAGGCACAGGCAGGCGAUCAGUGUGGCCAUUAGAAAGACUUUU ........((((((((((.((((((((((...(....).)))))))))).)((((((..(......)..)))))).(((.((........))..)))..)))))))))............ ( -40.40) >DroMoj_CAF1 5203 120 + 1 UAGAAGACAAUGGCCACGGUCUGAGUGGUGGUGGUGACCACGACCCAGGCGGAUGAGAAGAAGAUGCCGCUGAUCAUGAGUGAGAGGCAGGCAAUCAAGGUGGCCAGCAGAAAGAUGCGU ..........(((((((.(((((.((.(((((....))))).)).)))))............(((((((((..(((....)))..))).))).)))...)))))))(((......))).. ( -48.40) >DroAna_CAF1 1599 120 + 1 UAGAAAACUAUAGCCACCACCUGGGUGGUGGUGGUCACCACAAUCCAGGCGGAGGAGAACAGAAUCCCGCUGGCCAUCAAUGAGACACAGGCGAUCAAGGUGGCCAGGAGGAAGACCCUC .............((.((.(((((((.(((((....))))).))))))).)).)).........(((..(((((((((..(((...........))).)))))))))..)))........ ( -46.30) >consensus UAGAAGACAAUAGCCACCACCUGAGUGGUGGUGGUGACCACCACCCAGGCGGAUGAGAAGAAUAUGCCGCUGAUCAUCAGCGAUAGGCAGGCAAUCAGCGUGGCCAGCAGAAAGACCCGU ............(((((..((((.((.(((((....))))).)).))))..................(((((.....))))).................)))))................ (-20.03 = -20.07 + 0.03)

| Location | 19,044,789 – 19,044,909 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.17 |
| Mean single sequence MFE | -40.47 |
| Consensus MFE | -21.19 |
| Energy contribution | -22.11 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19044789 120 - 22224390 ACGGGUCUUCCUCCUGGCCACGCUCAUUGCCUGCGUAUCGCUGAUGGUCAGCGGGAUCCUGUUCUCCUCCGCCUGGAUCGUGGUCACAACCACCACACAGGUGGUAGCUAUUGUCUUCUA (((((...((((.(((((((.((.....))..(((...)))...))))))).)))).)))))....(((((((((..(.(((((....))))).)..))))))).))............. ( -44.30) >DroVir_CAF1 5488 120 - 1 GCGCAUCUUUCUGCUAGCCACACUGAUUGCAUGCCUAUCGCUGAUGAUCAGUGGCAUAUUUUUCUCAUCCGCCUGGGUGGUGGUCACCACCACCACUCAAACGGUGGCUAUUGUCUUCUA ..(((......)))(((((((((((((((((.((.....)))).)))))))))...............(((..(((((((((((....)))))))))))..))))))))).......... ( -44.80) >DroGri_CAF1 6274 120 - 1 ACGAAUGUUUCUGCUGGCCACGCUGAUUGCAUGUCUAUCGCUGAUGAUCAGCGGCAUAUUCUUCUCAUCCGCCUGGGUGGUUGUCACCACCACCACUCAAACCGUUGCGAUUGUCUUCUA ..((((((..((((((..((((.((((.........)))).)).))..)))))).))))))....((..(((.((((((((.((....)).)))))))).......)))..))....... ( -30.50) >DroWil_CAF1 1489 120 - 1 AAAAGUCUUUCUAAUGGCCACACUGAUCGCCUGCCUGUGCCUCAUGAUAAGCGGUAUACUCUUCUCAUCCGCCUGGGUGAUAGUCACCACAACCACCCAAGUGGUGGCCAUUGUCUUUUA (((((......((((((((((...(((.....(..((((((.(.......).))))))..).....)))(((.((((((...((....))...)))))).))))))))))))).))))). ( -33.90) >DroMoj_CAF1 5203 120 - 1 ACGCAUCUUUCUGCUGGCCACCUUGAUUGCCUGCCUCUCACUCAUGAUCAGCGGCAUCUUCUUCUCAUCCGCCUGGGUCGUGGUCACCACCACCACUCAGACCGUGGCCAUUGUCUUCUA ..(((......)))(((((((...(((.(((.((...(((....)))...))))))))..............((((((.(((((....))))).))))))...))))))).......... ( -37.60) >DroAna_CAF1 1599 120 - 1 GAGGGUCUUCCUCCUGGCCACCUUGAUCGCCUGUGUCUCAUUGAUGGCCAGCGGGAUUCUGUUCUCCUCCGCCUGGAUUGUGGUGACCACCACCACCCAGGUGGUGGCUAUAGUUUUCUA ((((.....))))((((((((..(((.((....))..)))..).))))))).((((..((((...((.((((((((...((((((.....)))))))))))))).))..))))..)))). ( -51.70) >consensus ACGAAUCUUUCUGCUGGCCACACUGAUUGCCUGCCUAUCGCUGAUGAUCAGCGGCAUACUCUUCUCAUCCGCCUGGGUGGUGGUCACCACCACCACUCAAACGGUGGCUAUUGUCUUCUA ..............(((((((.............................((((..............)))).(((((.(((((....))))).)))))....))))))).......... (-21.19 = -22.11 + 0.92)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:56 2006