
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,040,301 – 19,040,472 |
| Length | 171 |
| Max. P | 0.963096 |

| Location | 19,040,301 – 19,040,392 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 73.93 |
| Mean single sequence MFE | -26.00 |
| Consensus MFE | -16.94 |
| Energy contribution | -19.07 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827404 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
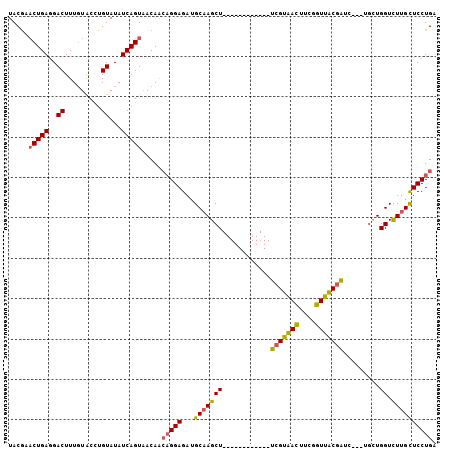
>X_DroMel_CAF1 19040301 91 + 22224390 UACGGACUGAGGACUUUGUGCCUGUAUAUCAGUAACAGAAGGAGAUGCAAGCU------------UCGUAACUUCGGUUACGAUC---UGCUGGUUUUGCUCCUGA ..............(((((..(((.....)))..)))))(((((..((.(((.------------(((((((....)))))))..---.))).))....))))).. ( -29.80) >DroSec_CAF1 9069 94 + 1 UACGCACUGAGGACUUUGUACGUGUAUAUCAGUAACAACAGGAGAUGCAAGCU------------UCGUAACUUCGGUUACGAUAUUAUCCUGGUCUUGUUCCUGA .....(((((..((.........))...))))).....(((((...(((((((------------(((((((....))))))).........)).)))))))))). ( -27.00) >DroYak_CAF1 8714 103 + 1 UACAAUCUGAUGACUUUCUACCUGUAUAUCAGUUACAACCGGAGAUGCUAGCUGAAAACUUUCUUUUGGGAUUUCGGUUUCUGUC---UGUUGGUCUUGCUCCUGA ......((((((((.........)).)))))).....((((((((((..((((((((.(((......))).))))))))..))))---).)))))........... ( -21.20) >consensus UACGAACUGAGGACUUUGUACCUGUAUAUCAGUAACAACAGGAGAUGCAAGCU____________UCGUAACUUCGGUUACGAUC___UGCUGGUCUUGCUCCUGA .....(((((..((.........))...))))).....(((((...(((((((............(((((((....))))))).........)).)))))))))). (-16.94 = -19.07 + 2.12)


| Location | 19,040,327 – 19,040,432 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.62 |
| Mean single sequence MFE | -30.33 |
| Consensus MFE | -18.17 |
| Energy contribution | -18.63 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19040327 105 + 22224390 UAUCAGUAACAGAAGGAGAUGCAAGCU------------UCGUAACUUCGGUUACGAUC---UGCUGGUUUUGCUCCUGAAUCUCAGCAGAAAACGAGAGAUUUUCCCAUUGGAUUUCGG ..(((((......(((((..((.(((.------------(((((((....)))))))..---.))).))....)))))(((((((............)))))))....)))))....... ( -30.90) >DroSec_CAF1 9095 108 + 1 UAUCAGUAACAACAGGAGAUGCAAGCU------------UCGUAACUUCGGUUACGAUAUUAUCCUGGUCUUGUUCCUGAAUCUCAGCAGGAUUUGGGAGAUUUUCCCAUUGGAUUUCGG ...............(((((.(((...------------(((((((....)))))))........(((.....((((..(((((.....)))))..))))......)))))).))))).. ( -31.80) >DroYak_CAF1 8740 117 + 1 UAUCAGUUACAACCGGAGAUGCUAGCUGAAAACUUUCUUUUGGGAUUUCGGUUUCUGUC---UGUUGGUCUUGCUCCUGAAUUUCAGCUUCAAACGGGAGAUUUUUCUCUUGGAUUUCAA ..((((...((((((((((((..((((((((.(((......))).))))))))..))))---).)))))..))...))))..............(((((((....)))))))........ ( -28.30) >consensus UAUCAGUAACAACAGGAGAUGCAAGCU____________UCGUAACUUCGGUUACGAUC___UGCUGGUCUUGCUCCUGAAUCUCAGCAGAAAACGGGAGAUUUUCCCAUUGGAUUUCGG .........(((..(((((.(((((((............(((((((....))))))).........)).)))))))..(((((((............))))))))))..)))........ (-18.17 = -18.63 + 0.46)


| Location | 19,040,355 – 19,040,472 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 65.55 |
| Mean single sequence MFE | -33.70 |
| Consensus MFE | -17.10 |
| Energy contribution | -20.33 |
| Covariance contribution | 3.24 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19040355 117 + 22224390 CGUAACUUCGGUUACGAUC---UGCUGGUUUUGCUCCUGAAUCUCAGCAGAAAACGAGAGAUUUUCCCAUUGGAUUUCGGAGACAGCCGAGUAAUCCUCCUGGGUCUCGGUUAUAGUCUG ((((((....)))))).((---((((((..((.(....)))..))))))))...(((((.....(((....)))))))).(((((((((((..((((....)))))))))))...)))). ( -40.70) >DroSec_CAF1 9123 120 + 1 CGUAACUUCGGUUACGAUAUUAUCCUGGUCUUGUUCCUGAAUCUCAGCAGGAUUUGGGAGAUUUUCCCAUUGGAUUUCGGAGACAAUCGGGUAAUCCUCCUGGAUCUCGGUUAUAGUUUG ((((((....))))))...........(((((..(((..(((((.....)))))..)))((...(((....)))..)).)))))(((((((..((((....)))))))))))........ ( -38.90) >DroYak_CAF1 8780 89 + 1 UGGGAUUUCGGUUUCUGUC---UGUUGGUCUUGCUCCUGAAUUUCAGCUUCAAACGGGAGAUUUUUCUCUUGGAUUUCAAAGACAAUCGGGC---------------------------- .(..((....))..).(((---((...(((((....(((.....))).......(((((((....))))))).......)))))...)))))---------------------------- ( -21.50) >consensus CGUAACUUCGGUUACGAUC___UGCUGGUCUUGCUCCUGAAUCUCAGCAGAAAACGGGAGAUUUUCCCAUUGGAUUUCGGAGACAAUCGGGUAAUCCUCCUGG_UCUCGGUUAUAGU_UG ((((((....))))))...........(((((....(((.....)))..........(((((((.......))))))).)))))(((((((..((((....)))))))))))........ (-17.10 = -20.33 + 3.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:48 2006