
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,028,870 – 19,028,975 |
| Length | 105 |
| Max. P | 0.571603 |

| Location | 19,028,870 – 19,028,975 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 81.31 |
| Mean single sequence MFE | -33.48 |
| Consensus MFE | -25.62 |
| Energy contribution | -26.07 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
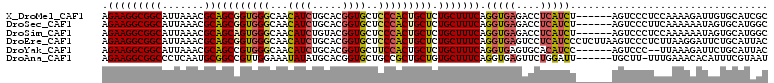
>X_DroMel_CAF1 19028870 105 + 22224390 AGAAGGCGGCAUUAAACGCAGCGGUGGGCAACAUCUGCACGGUGCUCCCACUGCUCUGCUUUCAGGUGAGACCUCAUCU------AGUCCCUCCAAAAGAUUGUGCAUCGC .((((((((..........(((((((((...((((.....))))..)))))))))))))))))((((((....))))))------.......................... ( -33.40) >DroSec_CAF1 2459 105 + 1 AGAAGGCGGCAUUAAACGCAGCGGUGGGCAACAUCUGCACGGUGCUCCCACUGCUCUGCUUUCAGGUGAGACCUCAUCU------AGUCCCUUCAAAAAAUAGUGCAUGGC .(((((.(((.......(((((((((((...((((.....))))..))))))))..)))....((((((....))))))------.))))))))................. ( -37.00) >DroSim_CAF1 2445 105 + 1 AGAAGGCGGCAUUAAACGCAGCAGUGGGCAACAUCUGUACGGUGCUCCCACUGCUCUGCUUUCAGGUGAGACCUCAUCU------AGUCCCUCCAAAAAAUAGUGCAUGGC .((((((((..........(((((((((...((((.....))))..)))))))))))))))))((((((....))))))------.......................... ( -34.10) >DroEre_CAF1 2866 111 + 1 AGAAGGCGGCAUUAAACGCAGCGGUGGGCAACAUCUGCACGGUGCUCCCACUGCUCUGCUUUCAGGUGAGUCCUCAUCCCUCUUAAGUCCCUCUUAAGGAUUCUGCAUUAC .((((((((..........(((((((((...((((.....))))..))))))))))))))))).(((((....)))))..(((((((.....)))))))............ ( -36.90) >DroYak_CAF1 2462 103 + 1 AGAAGGCGGCAUUAAACGCAGCCGUGGGCAACAUCUGCACGGUGCUUCCACUGCUCUGCUUUCAGGUGAGUGCACAUCC------AGUCCC--UUAAAGAUUCUGCAUUAC .(((((((........))).(((((((....).....)))))).))))...(((....((((.(((.((.((......)------).))))--).)))).....))).... ( -27.80) >DroAna_CAF1 2188 104 + 1 AGAAGGCGGCCCUCAAUGCGGCCGUUGGAAAUAUAUGCACGGUGCUGCCGCUGCUGUGCUUUCAGGUGAGUUCUGGAUU------UGCUU-UUUGAAACACAUUUCGUAAU ....((((((((.....).))))))).(((((....((((((.....))).))).(((.(((((((..((((...))))------..)).-..)))))))))))))..... ( -31.70) >consensus AGAAGGCGGCAUUAAACGCAGCGGUGGGCAACAUCUGCACGGUGCUCCCACUGCUCUGCUUUCAGGUGAGACCUCAUCU______AGUCCCUCCAAAAGAUAGUGCAUGAC .(((((((((.......))(((((((((...((((.....))))..))))))))).))))))).(((((....)))))................................. (-25.62 = -26.07 + 0.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:44 2006