| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,012,088 – 19,012,206 |
| Length | 118 |
| Max. P | 0.730663 |

| Location | 19,012,088 – 19,012,206 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.51 |
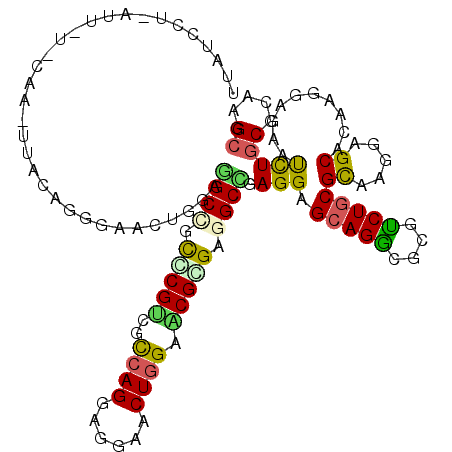
| Mean single sequence MFE | -42.02 |
| Consensus MFE | -28.60 |
| Energy contribution | -27.88 |
| Covariance contribution | -0.72 |
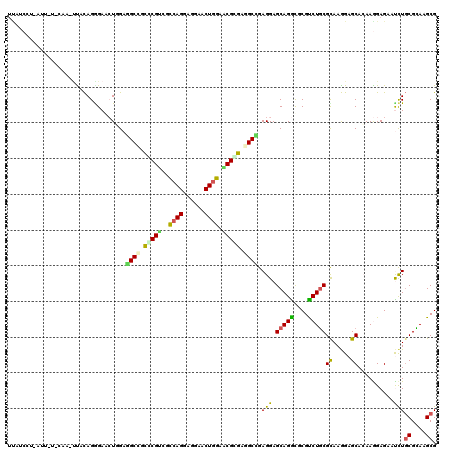
| Combinations/Pair | 1.56 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
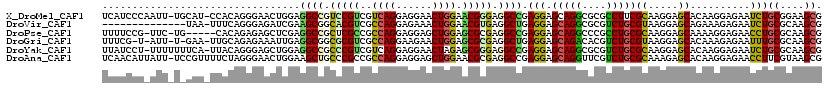
>X_DroMel_CAF1 19012088 118 - 22224390 UCAUCCCAAUU-UGCAU-CCACAGGGAACUGGAGGCCGUCCGUCGUCAGGAGGAACUGGAACGGGAGGCCGAGGAGCAGGCGCGCCUUCGCAAGGAGCACAAGGAGAAUCUGCGGAAGCG ...((((..((-(((.(-((.(((....)))..((((..((((..((((......)))).))))..))))..)))))))).(.((.(((....))))).)..)).))....((....)). ( -47.50) >DroVir_CAF1 894 105 - 1 --------------UAA-UUUCAGGGAGAUCGAAGCGGCACGUCGCCAGGAGAAACUGGAACGUGAGGCUGAGGAGCAGGCGCGUCUGCGUAAGGAGCAGAAAGAGAAUCUGCGCAAGCG --------------...-...........((..(((..(((((..((((......)))).)))))..)))..)).((..((((.(((((.......))))).((.....))))))..)). ( -36.00) >DroPse_CAF1 1506 113 - 1 UUUUCCG-UUC-UG-----CACAGAGAGCUCGAGGCCGCUCGCCGCCAGGAGGAGCUGGAGCGCGAGGCCGAGGAGCAGGCCCGCCUGCGCAAGGAGCAAAAGGAGAACCUGCGCAAGCG ((((((.-(((-(.-----...)))).((((..((((..(((((.((((......)))).).)))))))).....(((((....))))).....))))....))))))...((....)). ( -48.10) >DroGri_CAF1 880 115 - 1 UUUCG-U-AUU-U-GAA-UUGCAGAGAAAUUGAGGCGGCGCGUCGCCAGGAAGAACUGGAGCGCGAGGCUGAGGAGCAGACACGUCUGCGUAAGGAGCACAAAGAGAAUUUGCGCAAGCG .(((.-.-.((-(-(..-.(((...........(((..(((((..((((......)))).)))))..))).....(((((....))))).......)))))))..)))...((....)). ( -37.30) >DroYak_CAF1 872 118 - 1 UUAUCCU-UUUUUUUCA-UUACAGGGAGCUGGAGGCCGCCCGUCGUCAGGAGGAACUAGAGCGGGAGGCCGAGGAGCAGGCGCGUCUGCGCAAGGAGCACAAGGAGAAUCUGCGCAAGCG ...((((-(........-...(((....)))..((((.(((((..((((......)).))))))).)))))))))((..((((((((.(....).)).))..((.....))))))..)). ( -43.70) >DroAna_CAF1 890 119 - 1 UCAACAUUAUU-UCCGUUUUCUAGGGAACUGGAAGCUGCCCGCCGCCAGGAGGAGCUGGAACGCGAGGCCGAGGAGCAGGUUCGUCUGCGCAAAGAGCACAAGGAGAACCUUCGUAAGCG ...((......-(((..(((((((....)))))))(.((((((..((((......))))...))).))).).)))(.((((((..(((.((.....)).).))..)))))).)))..... ( -39.50) >consensus UUAUCCU_AUU_U_CAA_UUACAGGGAACUGGAGGCCGCCCGUCGCCAGGAGGAACUGGAACGCGAGGCCGAGGAGCAGGCGCGUCUGCGCAAGGAGCACAAGGAGAAUCUGCGCAAGCG .................................((((.(((((..((((......)))).))))).)))).(((.(((((....)))))((.....))..........)))((....)). (-28.60 = -27.88 + -0.72)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:38 2006