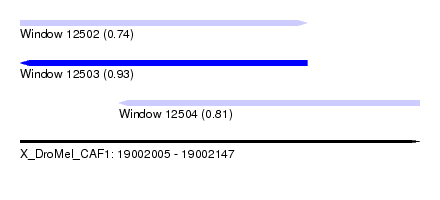
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,002,005 – 19,002,147 |
| Length | 142 |
| Max. P | 0.933106 |

| Location | 19,002,005 – 19,002,107 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.01 |
| Mean single sequence MFE | -38.67 |
| Consensus MFE | -17.83 |
| Energy contribution | -21.19 |
| Covariance contribution | 3.36 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
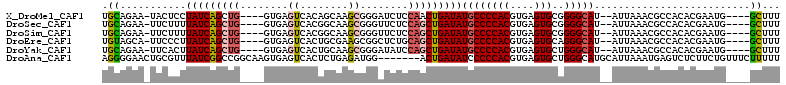
>X_DroMel_CAF1 19002005 102 + 22224390 UGCAGAA-UACUCCUAUCAGCUG----GUGAGUCACAGCAAGCGGGAUCUCCAACUGAUAUGCCCCACGUGAGUGCGGGGCAU--AUUAAACGCCACACGAAUG----GCUUU ((.(((.-...(((..(..((((----........))))..)..)))))).))..(((((((((((((....))..)))))))--))))...((((......))----))... ( -35.70) >DroSec_CAF1 24362 102 + 1 UGCAGAA-UUCUUUUAUCAGCUG----GUGAGUCACGGCAAGCGGGUUCUCCAGCUGAUAUGCCCCACGUGAGUGCGGGGCAU--AUUAAACGCCACACGAAUG----GCUUU .......-........(((((((----(.((..(.((.....)))..)).))))))))((((((((((....))..)))))))--)......((((......))----))... ( -42.10) >DroSim_CAF1 18948 102 + 1 UGCAGAA-UUCUUUUAUCAGCUG----GUGAGUCACGGCAAGCGGGUUCUCCAGCUGAUAUGCCCCACGUGAGUGCGGGGCAU--AUUAAACGCCACACGAAUG----GCUUU .......-........(((((((----(.((..(.((.....)))..)).))))))))((((((((((....))..)))))))--)......((((......))----))... ( -42.10) >DroEre_CAF1 23542 102 + 1 UGUAGCA-UUCCCUUAUCAGCUG----GUGAGUCACUGCGAAGCGGCUCUGCAGCUGAUAUGCCCCACGUGAGUGCAGGGCAU--AUUAAACGCCACACGAAUG----GCUUU .......-........(((((((----..(((((.((....)).)))))..)))))))((((((((((....)))..))))))--)......((((......))----))... ( -41.20) >DroYak_CAF1 24682 102 + 1 UGCAGAA-UUCACUUAUCAGCUG----GUGAGUCACUGCAAGCGGGAUAUCCAGCUGAUAUGCCCCACGUGAGUGCUGGGCAU--AUUAAACGCCACACGAAUG----GCUUU (((((.(-((((((........)----))))))..)))))(((.((....)).)))((((((((((((....)))..))))))--)))....((((......))----))... ( -41.90) >DroAna_CAF1 28997 106 + 1 AGGGGAACUGCGUUUAUCGGCCGGCAAGUGAGUCACUCUGAGAUGG-------ACUGAUAUCCCCCACGUGAGUGCUGGGCAUGCAUUAAAUGAGUCUCUUCUGUUUCUUUUU (((((((((.(((((((((((((.((((((...)))).))...)))-------.))))((((((.(((....)))..))).)))...))))))))).)))))).......... ( -29.00) >consensus UGCAGAA_UUCUCUUAUCAGCUG____GUGAGUCACUGCAAGCGGGAUCUCCAGCUGAUAUGCCCCACGUGAGUGCGGGGCAU__AUUAAACGCCACACGAAUG____GCUUU .((...........(((((((((........((........))........)))))))))((((((((....)))..)))))..........................))... (-17.83 = -21.19 + 3.36)
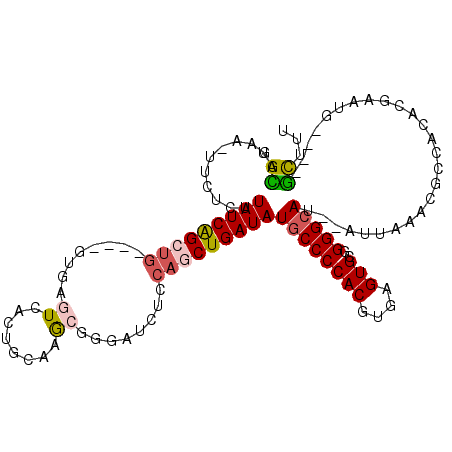
| Location | 19,002,005 – 19,002,107 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 81.01 |
| Mean single sequence MFE | -37.85 |
| Consensus MFE | -21.58 |
| Energy contribution | -22.39 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933106 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19002005 102 - 22224390 AAAGC----CAUUCGUGUGGCGUUUAAU--AUGCCCCGCACUCACGUGGGGCAUAUCAGUUGGAGAUCCCGCUUGCUGUGACUCAC----CAGCUGAUAGGAGUA-UUCUGCA ...((----(((....)))))((..(((--(((((((((......))))))).((((((((((.((...(((.....)))..)).)----)))))))))...)))-))..)). ( -43.20) >DroSec_CAF1 24362 102 - 1 AAAGC----CAUUCGUGUGGCGUUUAAU--AUGCCCCGCACUCACGUGGGGCAUAUCAGCUGGAGAACCCGCUUGCCGUGACUCAC----CAGCUGAUAAAAGAA-UUCUGCA ...((----(((....)))))((..(((--.((((((((......))))))))((((((((((.((...(((.....)))..)).)----))))))))).....)-))..)). ( -43.10) >DroSim_CAF1 18948 102 - 1 AAAGC----CAUUCGUGUGGCGUUUAAU--AUGCCCCGCACUCACGUGGGGCAUAUCAGCUGGAGAACCCGCUUGCCGUGACUCAC----CAGCUGAUAAAAGAA-UUCUGCA ...((----(((....)))))((..(((--.((((((((......))))))))((((((((((.((...(((.....)))..)).)----))))))))).....)-))..)). ( -43.10) >DroEre_CAF1 23542 102 - 1 AAAGC----CAUUCGUGUGGCGUUUAAU--AUGCCCUGCACUCACGUGGGGCAUAUCAGCUGCAGAGCCGCUUCGCAGUGACUCAC----CAGCUGAUAAGGGAA-UGCUACA .....----......((((((((((...--.(((((..(......)..)))))(((((((((..(((.((((....)))).)))..----)))))))))...)))-))))))) ( -44.60) >DroYak_CAF1 24682 102 - 1 AAAGC----CAUUCGUGUGGCGUUUAAU--AUGCCCAGCACUCACGUGGGGCAUAUCAGCUGGAUAUCCCGCUUGCAGUGACUCAC----CAGCUGAUAAGUGAA-UUCUGCA ...((----(((....)))))((..(((--.(((((.((......)).)))))((((((((((......(((.....))).....)----))))))))).....)-))..)). ( -35.50) >DroAna_CAF1 28997 106 - 1 AAAAAGAAACAGAAGAGACUCAUUUAAUGCAUGCCCAGCACUCACGUGGGGGAUAUCAGU-------CCAUCUCAGAGUGACUCACUUGCCGGCCGAUAAACGCAGUUCCCCU ...........................(((...(((..(((....))).))).((((.((-------(.......(((((...)))))...))).))))...)))........ ( -17.60) >consensus AAAGC____CAUUCGUGUGGCGUUUAAU__AUGCCCCGCACUCACGUGGGGCAUAUCAGCUGGAGAACCCGCUUGCAGUGACUCAC____CAGCUGAUAAAAGAA_UUCUGCA ...............(((((...........((((((((......))))))))(((((((((............................))))))))).........))))) (-21.58 = -22.39 + 0.81)

| Location | 19,002,040 – 19,002,147 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 80.48 |
| Mean single sequence MFE | -33.73 |
| Consensus MFE | -15.99 |
| Energy contribution | -18.27 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19002040 107 - 22224390 ACUGGCACUCGUUCAGGUGAAUGAAUGUAAAUAUUAUUCAAAAGCCAUUCGUGUGGCGUUUAAUAUGC--CCCGCACU-----CACGUGGGGCAUAUCAGUUGGAGAUCCCGCU (((((((.((((((....)))))).)))...............(((((....))))).....((((((--(((((...-----...))))))))))))))).((.....))... ( -37.60) >DroPse_CAF1 24613 111 - 1 UCCCGAACACACUCAUGUGAAUAAAUGUAAAUAUUAUUCAAAACUCAUUCGUGUGGCAUUUUAUAGGCACCCUGCACAUGGUCUUUGUCGGGCAUAUCAGCACACGCACA---C .(((((....((.((((((......(((..(((..((.(...((......))...).))..)))..))).....)))))))).....))))).......((....))...---. ( -20.70) >DroSec_CAF1 24397 107 - 1 ACUGGCACUCGUUCAGCUGAAUGAAUGUAAAUAUUAUUCAAAAGCCAUUCGUGUGGCGUUUAAUAUGC--CCCGCACU-----CACGUGGGGCAUAUCAGCUGGAGAACCCGCU ...((...((.(((((((((.((((((.......))))))...(((((....)))))......(((((--(((((...-----...)))))))))))))))))))))..))... ( -44.10) >DroSim_CAF1 18983 107 - 1 ACUGGCACUCGUUCAGGUGAAUGAAUGUAAAUAUUAUUCAAAAGCCAUUCGUGUGGCGUUUAAUAUGC--CCCGCACU-----CACGUGGGGCAUAUCAGCUGGAGAACCCGCU ...((...((.(((((.(((.((((((.......))))))...(((((....)))))......(((((--(((((...-----...))))))))))))).)))))))..))... ( -39.60) >DroEre_CAF1 23577 107 - 1 ACUCGCACUCGUUCAGGUGAAUGAAUGUAAAUAUUAUUCAAAAGCCAUUCGUGUGGCGUUUAAUAUGC--CCUGCACU-----CACGUGGGGCAUAUCAGCUGCAGAGCCGCUU ....((.(((...(((.(((.((((((.......))))))...(((((....)))))......(((((--((..(...-----...)..)))))))))).)))..)))..)).. ( -37.80) >DroPer_CAF1 26818 111 - 1 UCCCGAACACACUCAUGUGAAUAAAUGUAAAUAUUAUUCAAAACUCAUUCGUGUGGCAUUUUAUAGGCACCCUGCACAAUGUCUUCGUCGGGCAUAUCAGCACACGCACA---U .(((((.(((((..((((((((((((....)).))))))).....)))..)))))(((((.(.((((...)))).).))))).....))))).......((....))...---. ( -22.60) >consensus ACUCGCACUCGUUCAGGUGAAUGAAUGUAAAUAUUAUUCAAAAGCCAUUCGUGUGGCGUUUAAUAUGC__CCCGCACU_____CACGUGGGGCAUAUCAGCUGGAGAACCCGCU ...........(((((.(((.((((((.......))))))...(((((....)))))......(((((..(((((...........))))))))))))).)))))......... (-15.99 = -18.27 + 2.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:30 2006