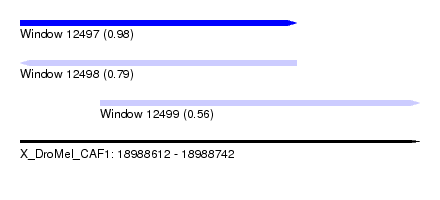
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,988,612 – 18,988,742 |
| Length | 130 |
| Max. P | 0.975800 |

| Location | 18,988,612 – 18,988,702 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 77.60 |
| Mean single sequence MFE | -40.46 |
| Consensus MFE | -24.13 |
| Energy contribution | -27.37 |
| Covariance contribution | 3.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18988612 90 + 22224390 ----GAAAUCGUCGAAAGGGGCGGUGGCGGUG------GGGGAGGCGGUGUACUCACCACGCCCCCGCUCACGAUGCACUGGCAGAAAUGGCAGUAGCGU ----...((((((....)((((((.((((.((------(..(((........))).))))))).)))))))))))((((((.((....)).)))).)).. ( -44.10) >DroSec_CAF1 11090 78 + 1 ----GAAAUCGGCGAAAGUGGCGGUGGC------------------GGUGUACUCACCACGCCCCCGCUCACGAUGCACUGGCAGAAAUGGCAGUAGCGU ----...((((((....))(((((.(((------------------((((....))))..))).)))))..))))((((((.((....)).)))).)).. ( -35.70) >DroSim_CAF1 13555 90 + 1 ----GAAAUCGGAGAAUGGGGCGGUGGCGGUG------GGGGAGGCGGUGUACUCACCACGCCCCCGCUCACGAUGCACUGGCAGAAAUGGCAGUAGCGU ----...((((.......((((((.((((.((------(..(((........))).))))))).)))))).))))((((((.((....)).)))).)).. ( -40.80) >DroEre_CAF1 11055 96 + 1 ----GAAAUGGGCAAAAGGGGCGGUGGCGUUGGGGGUGGGGGAGGCGGUGUACUCACCACGCCCCCGCUCACGAUGCACUGGCAGAAAUGGCAGUAGCGU ----................((....((((((.(((((((((..(.((((....)))).).))))))))).))))))((((.((....)).)))).)).. ( -45.00) >DroYak_CAF1 11627 96 + 1 ----GAAGUCGGCGAAAGGGGCGGUGGCGGUGGGGGAGGGGGAGGCGGUGUACUCACCACGCCCCCGCUCACGAUGCACUGACAGAAAUGGCAGUAGCGU ----...((((.(....)((((((.((((.(((..(((..............))).))))))).)))))).))))((((((.((....)).)))).)).. ( -40.44) >DroMoj_CAF1 24141 91 + 1 GCUU---GACGCAGACUGCGGCUGAGGCUGUU------GUUGCUGCAGCGCUGCGACAACGCUGCCGCUCACAAUGCACUGGCAGAAGUGGCAAUAGCGA ..((---(.(((((.(((((((...(((....------))))))))))..))))).)))((((((((((.....(((....)))..))))))...)))). ( -36.70) >consensus ____GAAAUCGGCGAAAGGGGCGGUGGCGGUG______GGGGAGGCGGUGUACUCACCACGCCCCCGCUCACGAUGCACUGGCAGAAAUGGCAGUAGCGU ..................((((((.((((.........(......)((((....)))).)))).))))))...((((((((.((....)).)))).)))) (-24.13 = -27.37 + 3.24)


| Location | 18,988,612 – 18,988,702 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 77.60 |
| Mean single sequence MFE | -31.66 |
| Consensus MFE | -20.49 |
| Energy contribution | -20.99 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18988612 90 - 22224390 ACGCUACUGCCAUUUCUGCCAGUGCAUCGUGAGCGGGGGCGUGGUGAGUACACCGCCUCCCC------CACCGCCACCGCCCCUUUCGACGAUUUC---- ..((.((((.((....)).))))))((((((((.((((((((((((.((.............------.))))))).)))))))))).)))))...---- ( -33.14) >DroSec_CAF1 11090 78 - 1 ACGCUACUGCCAUUUCUGCCAGUGCAUCGUGAGCGGGGGCGUGGUGAGUACACC------------------GCCACCGCCACUUUCGCCGAUUUC---- ..((.((((.((....)).))))))...(((.((((.(((..((((....))))------------------))).))))))).............---- ( -27.50) >DroSim_CAF1 13555 90 - 1 ACGCUACUGCCAUUUCUGCCAGUGCAUCGUGAGCGGGGGCGUGGUGAGUACACCGCCUCCCC------CACCGCCACCGCCCCAUUCUCCGAUUUC---- ..((.((((.((....)).))))))((((.(((..(((((((((((.((.............------.))))))).))))))...)))))))...---- ( -33.64) >DroEre_CAF1 11055 96 - 1 ACGCUACUGCCAUUUCUGCCAGUGCAUCGUGAGCGGGGGCGUGGUGAGUACACCGCCUCCCCCACCCCCAACGCCACCGCCCCUUUUGCCCAUUUC---- ..((.((((.((....)).))))))...(((.(((((((.((((((....))))))..))))).........((....)).......)).)))...---- ( -29.90) >DroYak_CAF1 11627 96 - 1 ACGCUACUGCCAUUUCUGUCAGUGCAUCGUGAGCGGGGGCGUGGUGAGUACACCGCCUCCCCCUCCCCCACCGCCACCGCCCCUUUCGCCGACUUC---- ..((.((((.((....)).)))))).(((((((.((((((((((((.((....................))))))).))))))))))).)))....---- ( -31.65) >DroMoj_CAF1 24141 91 - 1 UCGCUAUUGCCACUUCUGCCAGUGCAUUGUGAGCGGCAGCGUUGUCGCAGCGCUGCAGCAAC------AACAGCCUCAGCCGCAGUCUGCGUC---AAGC .((((.(((((((...(((....)))..))).)))).))))(((.(((((.(((((.((...------..........)).)))))))))).)---)).. ( -34.12) >consensus ACGCUACUGCCAUUUCUGCCAGUGCAUCGUGAGCGGGGGCGUGGUGAGUACACCGCCUCCCC______CACCGCCACCGCCCCUUUCGCCGAUUUC____ .(((((((((.(((......))))))..)).))))(((((((((((.........................)))))).)))))................. (-20.49 = -20.99 + 0.50)

| Location | 18,988,638 – 18,988,742 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 84.20 |
| Mean single sequence MFE | -45.78 |
| Consensus MFE | -29.63 |
| Energy contribution | -33.38 |
| Covariance contribution | 3.75 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18988638 104 + 22224390 UG------GGGGAGGCGGUGUACUCACCACGCCCCCGCUCACGAUGCACUGGCAGAAAUGGCAGUAGCGUCCUUGGGCGGCAAACACGCCGUAGCCGCAACCCCGACAAA ..------((((..(((((.(((.............(((((.(((((((((.((....)).)))).)))))..)))))(((......)))))))))))..))))...... ( -48.30) >DroSec_CAF1 11114 94 + 1 ----------------GGUGUACUCACCACGCCCCCGCUCACGAUGCACUGGCAGAAAUGGCAGUAGCGUCCUUGGGCGGCAAACACGCCGUAGCCGCAACCCCGACAAA ----------------((((....))))..((....(((((.(((((((((.((....)).)))).)))))..)))))(((......)))......))............ ( -35.00) >DroSim_CAF1 13581 104 + 1 UG------GGGGAGGCGGUGUACUCACCACGCCCCCGCUCACGAUGCACUGGCAGAAAUGGCAGUAGCGUCCUUGGGCGGCAAACACGCCGUAGCCGCAACCCCGACAAA ..------((((..(((((.(((.............(((((.(((((((((.((....)).)))).)))))..)))))(((......)))))))))))..))))...... ( -48.30) >DroEre_CAF1 11081 110 + 1 UGGGGGUGGGGGAGGCGGUGUACUCACCACGCCCCCGCUCACGAUGCACUGGCAGAAAUGGCAGUAGCGUCCUUGGGCAGCAAACACGCCGUAGCCGCAUCCCCGACAAA .(((((((.((..(((((((....))))..((....(((((.(((((((((.((....)).)))).)))))..))))).))......)))....)).)))))))...... ( -52.50) >DroYak_CAF1 11653 110 + 1 UGGGGGAGGGGGAGGCGGUGUACUCACCACGCCCCCGCUCACGAUGCACUGACAGAAAUGGCAGUAGCGUCCUUGGGCGGCAAACACGCCAUAGCCGCAUCCGCGACAAA ..(.((((((((..(.((((....)))).).)))))(((((.(((((((((.((....)).)))).)))))..)))))(((......))).........))).)...... ( -48.30) >DroMoj_CAF1 24168 104 + 1 UU------GUUGCUGCAGCGCUGCGACAACGCUGCCGCUCACAAUGCACUGGCAGAAGUGGCAAUAGCGACCUUGUGCUGCAAAGACGCCAUAGCCGCAGCCGUGGCACA ..------(((..((((((((.(((....)))(((((((.....(((....)))..)))))))...........))))))))..)))((((..((....))..))))... ( -42.30) >consensus UG______GGGGAGGCGGUGUACUCACCACGCCCCCGCUCACGAUGCACUGGCAGAAAUGGCAGUAGCGUCCUUGGGCGGCAAACACGCCGUAGCCGCAACCCCGACAAA ........((((..(((((.................(((((.(((((((((.((....)).)))).)))))..)))))(((......)))...)))))..))))...... (-29.63 = -33.38 + 3.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:26 2006