
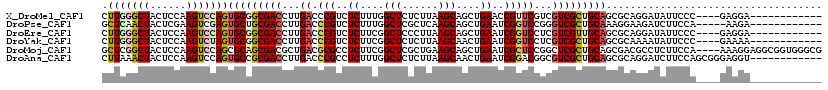
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,988,340 – 18,988,444 |
| Length | 104 |
| Max. P | 0.732514 |

| Location | 18,988,340 – 18,988,444 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.80 |
| Mean single sequence MFE | -38.92 |
| Consensus MFE | -25.42 |
| Energy contribution | -25.98 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
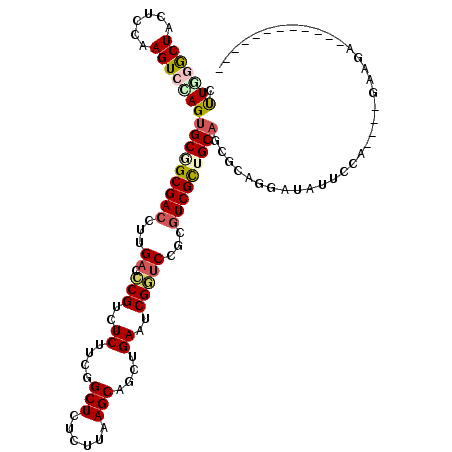
>X_DroMel_CAF1 18988340 104 - 22224390 CUUGGGCUACUCCAAGUCCAGUGCGGCGACCUUGACCCGUCUCUUUGGCUCUCUUAAGCAGCUGAACCGUUCGUCGUCGCUGCAGCGCAGGAUAUUCCC----GAGGA------------ ((((((....(((..((....(((((((((..(((..((..((....(((......)))....))..)).)))..)))))))))))...)))....)))----)))..------------ ( -35.10) >DroPse_CAF1 11690 103 - 1 GCUCAACUACUCGAAGUCGAGUGCUGCGACCUUGACCCGUCUCUUUGGCUCGCUCAAGCAGCUGAAUCGGUCCGGGUCGCUGCAAAGGAAGAUCUUCCA-----AAGA------------ ........(((((....)))))((.(((((((.((((.(....((..(((.((....)))))..)).))))).))))))).))...((((....)))).-----....------------ ( -40.40) >DroEre_CAF1 10774 104 - 1 CUUGGGCUACUCCAAGUCCAGUGCGGCGACCUUGACCCGUCUCUUCGGCUCCCUUAAGCAGCUGAAUCGGUCCUCGUCGUUGCAGCGCAGGAUAUUCCC----GAGGA------------ ((((((....(((..((....(((((((((...((((.(....(((((((.(.....).))))))).)))))...)))))))))))...)))....)))----)))..------------ ( -36.80) >DroYak_CAF1 11346 104 - 1 CUUGGGCUACUCCAAGUCUAGUGCGGCGACCUUGACCCGUCUCUUCGGCUCUCUUAAGCAACUGAAUCGGUCCUCGUCGCUGCAGCGCAAAAUAUUCCC----GAAAA------------ ((((((....)))))).....(((((((((...((((.(..((....(((......)))....))..)))))...)))))))))...............----.....------------ ( -30.70) >DroMoj_CAF1 23831 116 - 1 GCUCGGCUACUCCAAGUCCAGCGCAGCGACGCUGACGCGCCUCUUCGGCUCGCUGAAGCAGCUGAAUCGCUCCGGCUCGCUGCAGCGACGCCUCUUCCA----AAAGGAGGCGGUGGGCG (((.((((......)))).)))(((((((.((((..(((....(((((((.((....))))))))).)))..))))))))))).((..((((.(((((.----...))))).)))).)). ( -57.10) >DroAna_CAF1 10528 108 - 1 CUUAAACUACUCCAAGUCCAGUGCCGCGACCUUGACCCGCCUCUUUGGCUCUCUUAAGCAACUGAAUCGGACGGCGUCGCUGCAGCGCAGGAUCUUCCAGCGGGAGGU------------ .........((((........(((.(((((.(((..(((..((....(((......)))....))..))).))).))))).))).(((.((.....)).)))))))..------------ ( -33.40) >consensus CUUGGGCUACUCCAAGUCCAGUGCGGCGACCUUGACCCGUCUCUUCGGCUCUCUUAAGCAGCUGAAUCGGUCCGCGUCGCUGCAGCGCAGGAUAUUCCA____GAAGA____________ .(((((((......)))))))(((((((((...((.(((..((....(((......)))....))..)))))...))))))))).................................... (-25.42 = -25.98 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:22 2006