
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,953,864 – 18,954,164 |
| Length | 300 |
| Max. P | 0.999942 |

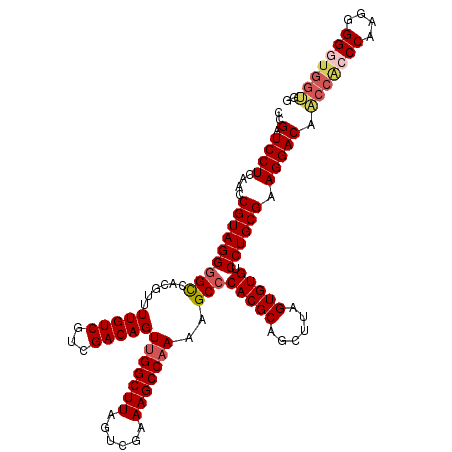
| Location | 18,953,864 – 18,953,979 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 91.04 |
| Mean single sequence MFE | -44.34 |
| Consensus MFE | -37.58 |
| Energy contribution | -38.66 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18953864 115 + 22224390 CCGAUCCUCAAGCGUAGGGGCCACGUUUUGUCGUCGACAGUUGGCUUAGUCGAAAGCCAAAAGCCCACGCAGCUUAGUGUGUCCUGCGAAGGACAACCACCCAAGGGGGUGGUGG (((.((((....(((((((((......(((((...)))))(((((((......)))))))..)))(((((......))))).)))))).))))).(((((((....))))))))) ( -45.80) >DroSec_CAF1 21798 115 + 1 CCGAUCCUCAACCGUAGGGGCCACGUUUUGUCGUCGACAGUUGGCUUAGUUGGAAGCCAAAAGCCCACGCAGCUUAGUGUGUCCUGCGAAGGACAACCACCCAGGGGGGUGGUGA ..(.((((....(((((((((......(((((...)))))(((((((......)))))))..)))(((((......))))).)))))).))))).(((((((....))))))).. ( -44.40) >DroSim_CAF1 10325 115 + 1 CCGAUCCUCAACCGUAGGGGCCACGUUUUGUCGUCGACAGUUGGCUUAGUUGAAAGCCAAAAGCCCACGCAGCUUAGUGUGUCCUGCGAAGGACAACCACCCAGGAGGGUGGUGG (((.((((....(((((((((......(((((...)))))(((((((......)))))))..)))(((((......))))).)))))).))))).(((((((....))))))))) ( -44.80) >DroEre_CAF1 22549 114 + 1 CCGAUCCUCAACCGUAGG-GUCACGUUUUGUCGUCGACAGUUGGCUUAGUCGAAAGCCAAAAGCCCACGCAGCUUAGUGUGUCCUGCGAAGGACACCCGCCCAAUGGGUUGGGUG ....((((....((((((-((((((......))).)))..(((((((......))))))).....(((((......))))).)))))).))))(((((((((...))).)))))) ( -43.00) >DroYak_CAF1 12076 109 + 1 CCGAUCCUCGGCCGUAGGGGCCACGUUUUGUCGUCGACAGUUGGCUUAGGCGAAAGCCCAAAGCCCACGCAGCUUAGUGUGUCCUGCGAAGGACACCCACCCAAUGGGU------ (((.....)))((((.(((((..(((.(((((...)))))..(((((.(((....)))..))))).)))..))...(.(((((((....))))))).).))).))))..------ ( -43.70) >consensus CCGAUCCUCAACCGUAGGGGCCACGUUUUGUCGUCGACAGUUGGCUUAGUCGAAAGCCAAAAGCCCACGCAGCUUAGUGUGUCCUGCGAAGGACAACCACCCAAGGGGGUGGUGG ..(.((((....(((((((((......(((((...)))))(((((((......)))))))..)))(((((......))))).)))))).))))).(((((((....))))))).. (-37.58 = -38.66 + 1.08)


| Location | 18,953,864 – 18,953,979 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 91.04 |
| Mean single sequence MFE | -44.04 |
| Consensus MFE | -37.38 |
| Energy contribution | -38.74 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18953864 115 - 22224390 CCACCACCCCCUUGGGUGGUUGUCCUUCGCAGGACACACUAAGCUGCGUGGGCUUUUGGCUUUCGACUAAGCCAACUGUCGACGACAAAACGUGGCCCCUACGCUUGAGGAUCGG (((((((((....)))))))((((((....))))))...((((((((((.(((..(((((((......)))))))..))).))).))....((((...))))))))).))..... ( -44.80) >DroSec_CAF1 21798 115 - 1 UCACCACCCCCCUGGGUGGUUGUCCUUCGCAGGACACACUAAGCUGCGUGGGCUUUUGGCUUCCAACUAAGCCAACUGUCGACGACAAAACGUGGCCCCUACGGUUGAGGAUCGG ..(((((((....))))))).((((((((.(((...(((.......(((.(((..(((((((......)))))))..))).))).......)))...))).))...))))))... ( -42.44) >DroSim_CAF1 10325 115 - 1 CCACCACCCUCCUGGGUGGUUGUCCUUCGCAGGACACACUAAGCUGCGUGGGCUUUUGGCUUUCAACUAAGCCAACUGUCGACGACAAAACGUGGCCCCUACGGUUGAGGAUCGG (((((((((....))))))).((((((((.(((...(((.......(((.(((..(((((((......)))))))..))).))).......)))...))).))...)))))).)) ( -45.14) >DroEre_CAF1 22549 114 - 1 CACCCAACCCAUUGGGCGGGUGUCCUUCGCAGGACACACUAAGCUGCGUGGGCUUUUGGCUUUCGACUAAGCCAACUGUCGACGACAAAACGUGAC-CCUACGGUUGAGGAUCGG ..(((((((....(((((((((((((....))))))).))....(((((.(((..(((((((......)))))))..))).))).))......).)-))...))))).))..... ( -44.00) >DroYak_CAF1 12076 109 - 1 ------ACCCAUUGGGUGGGUGUCCUUCGCAGGACACACUAAGCUGCGUGGGCUUUGGGCUUUCGCCUAAGCCAACUGUCGACGACAAAACGUGGCCCCUACGGCCGAGGAUCGG ------..((...(((((((((((((....))))))).))).((..(((.(((((.((((....)))))))))...((((...))))..)))..)))))...))(((.....))) ( -43.80) >consensus CCACCACCCCCUUGGGUGGUUGUCCUUCGCAGGACACACUAAGCUGCGUGGGCUUUUGGCUUUCGACUAAGCCAACUGUCGACGACAAAACGUGGCCCCUACGGUUGAGGAUCGG ..(((((((....))))))).((((((((.(((...(((.......(((.(((..(((((((......)))))))..))).))).......)))...))).))...))))))... (-37.38 = -38.74 + 1.36)



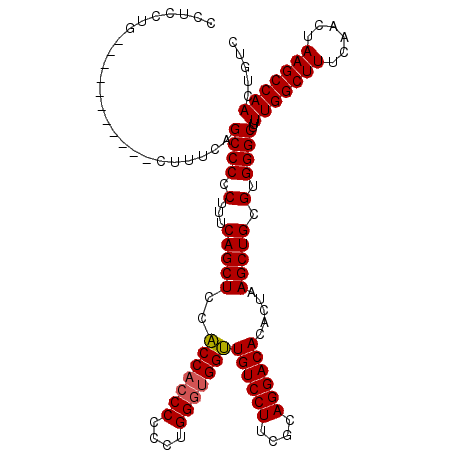
| Location | 18,953,899 – 18,954,006 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 87.11 |
| Mean single sequence MFE | -48.07 |
| Consensus MFE | -46.28 |
| Energy contribution | -46.02 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.09 |
| Mean z-score | -4.44 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.07 |
| SVM RNA-class probability | 0.998340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18953899 107 + 22224390 GACAGUUGGCUUAGUCGAAAGCCAAAAGCCCACGCAGCUUAGUGUGUCCUGCGAAGGACAACCACCCAAGGGGGUGGUGGAGCUGAAAGGGGGCGGAAAG-----------CAGGAGC .....(((((((......)))))))..((((.(.((((((....((((((....))))))(((((((....))))))).))))))...).))))......-----------....... ( -47.40) >DroSec_CAF1 21833 107 + 1 GACAGUUGGCUUAGUUGGAAGCCAAAAGCCCACGCAGCUUAGUGUGUCCUGCGAAGGACAACCACCCAGGGGGGUGGUGAAGCUGAAAGGGGGCUGAAAG-----------CAGGACG .....(((((((......))))))).(((((.(.((((((....((((((....))))))(((((((....))))))).))))))...).))))).....-----------....... ( -47.80) >DroSim_CAF1 10360 107 + 1 GACAGUUGGCUUAGUUGAAAGCCAAAAGCCCACGCAGCUUAGUGUGUCCUGCGAAGGACAACCACCCAGGAGGGUGGUGGAGCUGAAAGGGGGCUGAAAG-----------CAGGAGG .....(((((((......))))))).(((((.(.((((((....((((((....))))))(((((((....))))))).))))))...).))))).....-----------....... ( -48.20) >DroEre_CAF1 22583 117 + 1 GACAGUUGGCUUAGUCGAAAGCCAAAAGCCCACGCAGCUUAGUGUGUCCUGCGAAGGACACCCGCCCAAUGGGUUGGGUGAGCUGAAAGGGGGCUGAAAGAGGGCUGAAAGGGGGAA- ..((((((((((......)))))...(((((.(.((((((.(.(((((((....))))))).)((((((....))))))))))))...).))))).......)))))..........- ( -48.90) >consensus GACAGUUGGCUUAGUCGAAAGCCAAAAGCCCACGCAGCUUAGUGUGUCCUGCGAAGGACAACCACCCAAGGGGGUGGUGGAGCUGAAAGGGGGCUGAAAG___________CAGGAGG .....(((((((......))))))).(((((.(.((((((....((((((....))))))(((((((....))))))).))))))...).)))))....................... (-46.28 = -46.02 + -0.25)

| Location | 18,953,899 – 18,954,006 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 87.11 |
| Mean single sequence MFE | -40.65 |
| Consensus MFE | -38.47 |
| Energy contribution | -38.60 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.03 |
| Mean z-score | -4.38 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.05 |
| SVM RNA-class probability | 0.998275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18953899 107 - 22224390 GCUCCUG-----------CUUUCCGCCCCCUUUCAGCUCCACCACCCCCUUGGGUGGUUGUCCUUCGCAGGACACACUAAGCUGCGUGGGCUUUUGGCUUUCGACUAAGCCAACUGUC ((....)-----------).....((((.(...(((((..(((((((....)))))))((((((....)))))).....))))).).))))..(((((((......)))))))..... ( -40.90) >DroSec_CAF1 21833 107 - 1 CGUCCUG-----------CUUUCAGCCCCCUUUCAGCUUCACCACCCCCCUGGGUGGUUGUCCUUCGCAGGACACACUAAGCUGCGUGGGCUUUUGGCUUCCAACUAAGCCAACUGUC .......-----------.....(((((.(...((((((.(((((((....)))))))((((((....))))))....)))))).).))))).(((((((......)))))))..... ( -40.40) >DroSim_CAF1 10360 107 - 1 CCUCCUG-----------CUUUCAGCCCCCUUUCAGCUCCACCACCCUCCUGGGUGGUUGUCCUUCGCAGGACACACUAAGCUGCGUGGGCUUUUGGCUUUCAACUAAGCCAACUGUC .......-----------.....(((((.(...(((((..(((((((....)))))))((((((....)))))).....))))).).))))).(((((((......)))))))..... ( -41.60) >DroEre_CAF1 22583 117 - 1 -UUCCCCCUUUCAGCCCUCUUUCAGCCCCCUUUCAGCUCACCCAACCCAUUGGGCGGGUGUCCUUCGCAGGACACACUAAGCUGCGUGGGCUUUUGGCUUUCGACUAAGCCAACUGUC -..........(((.........(((((.(...((((((.(((((....))))).).(((((((....)))))))....))))).).))))).(((((((......)))))))))).. ( -39.70) >consensus CCUCCUG___________CUUUCAGCCCCCUUUCAGCUCCACCACCCCCCUGGGUGGUUGUCCUUCGCAGGACACACUAAGCUGCGUGGGCUUUUGGCUUUCAACUAAGCCAACUGUC ........................((((.(...(((((..(((((((....)))))))((((((....)))))).....))))).).))))..(((((((......)))))))..... (-38.47 = -38.60 + 0.13)

| Location | 18,953,939 – 18,954,046 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 85.19 |
| Mean single sequence MFE | -36.83 |
| Consensus MFE | -27.80 |
| Energy contribution | -28.05 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18953939 107 + 22224390 AGUGUGUCCUGCGAAGGACAACCACCCAAGGGGGUGGUGGAGCUGAAAGGGGGCGGAAAG-----------CAGGAGCGGUGGGUCUCUACGACUGCUGUGAAUACUUGGGCAGAAGU ....((((((....))))))(((((((....)))))))...(((..(((...((.....)-----------).(.((((((.(.......).)))))).).....))).)))...... ( -37.70) >DroSec_CAF1 21873 107 + 1 AGUGUGUCCUGCGAAGGACAACCACCCAGGGGGGUGGUGAAGCUGAAAGGGGGCUGAAAG-----------CAGGACGGGUGGGGCUCUACGACUGCUGUGAAUACUUGGGCAGAAGU ....((((((((........(((((((....)))))))..((((.......))))....)-----------))))))).((((....))))..(((((...........))))).... ( -37.50) >DroSim_CAF1 10400 107 + 1 AGUGUGUCCUGCGAAGGACAACCACCCAGGAGGGUGGUGGAGCUGAAAGGGGGCUGAAAG-----------CAGGAGGGGUGGGGCUCUACGACUGCUGUGAAUACUUGGGCAGAAGU ....((((((....))))))(((((((....)))))))....(((...(((........(-----------(((.((..((((....))))..)).)))).....)))...))).... ( -37.62) >DroEre_CAF1 22623 115 + 1 AGUGUGUCCUGCGAAGGACACCCGCCCAAUGGGUUGGGUGAGCUGAAAGGGGGCUGAAAGAGGGCUGAAAGGGGGAA---GGGGACUCUGCAACUGCUGUGAAUACUUGGGCAGAAGU ...(((((((....))))))).(((((((....)))))))..((....)).((((.......)))).....(.(((.---(....)))).)..(((((...........))))).... ( -34.50) >consensus AGUGUGUCCUGCGAAGGACAACCACCCAAGGGGGUGGUGGAGCUGAAAGGGGGCUGAAAG___________CAGGAGGGGUGGGGCUCUACGACUGCUGUGAAUACUUGGGCAGAAGU ....((((((....))))))(((((((....)))))))..((((.......))))..................((((........))))((..(((((...........)))))..)) (-27.80 = -28.05 + 0.25)


| Location | 18,953,939 – 18,954,046 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 85.19 |
| Mean single sequence MFE | -27.89 |
| Consensus MFE | -21.44 |
| Energy contribution | -21.38 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.920916 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18953939 107 - 22224390 ACUUCUGCCCAAGUAUUCACAGCAGUCGUAGAGACCCACCGCUCCUG-----------CUUUCCGCCCCCUUUCAGCUCCACCACCCCCUUGGGUGGUUGUCCUUCGCAGGACACACU ....(..((((((.......(((((..(..(.......)..)..)))-----------))....((.........))...........))))))..).((((((....)))))).... ( -25.90) >DroSec_CAF1 21873 107 - 1 ACUUCUGCCCAAGUAUUCACAGCAGUCGUAGAGCCCCACCCGUCCUG-----------CUUUCAGCCCCCUUUCAGCUUCACCACCCCCCUGGGUGGUUGUCCUUCGCAGGACACACU ((..((((.............))))..))............((((((-----------(....(((.........)))..(((((((....)))))))........)))))))..... ( -29.72) >DroSim_CAF1 10400 107 - 1 ACUUCUGCCCAAGUAUUCACAGCAGUCGUAGAGCCCCACCCCUCCUG-----------CUUUCAGCCCCCUUUCAGCUCCACCACCCUCCUGGGUGGUUGUCCUUCGCAGGACACACU ((..((((.............))))..)).((((..........(((-----------....)))..........)))).(((((((....)))))))((((((....)))))).... ( -28.07) >DroEre_CAF1 22623 115 - 1 ACUUCUGCCCAAGUAUUCACAGCAGUUGCAGAGUCCCC---UUCCCCCUUUCAGCCCUCUUUCAGCCCCCUUUCAGCUCACCCAACCCAUUGGGCGGGUGUCCUUCGCAGGACACACU ....((((((((...........(((((.((((.....---............................)))))))))...........))))))))(((((((....)))))))... ( -27.86) >consensus ACUUCUGCCCAAGUAUUCACAGCAGUCGUAGAGCCCCACCCCUCCUG___________CUUUCAGCCCCCUUUCAGCUCCACCACCCCCCUGGGUGGUUGUCCUUCGCAGGACACACU ..((((((....((.......))....))))))...............................................(((((((....)))))))((((((....)))))).... (-21.44 = -21.38 + -0.06)


| Location | 18,953,979 – 18,954,086 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.35 |
| Mean single sequence MFE | -29.50 |
| Consensus MFE | -20.16 |
| Energy contribution | -19.88 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18953979 107 + 22224390 AGCUGAAAGGGGGCGGAAAG-----------CAGGAGC--GGUGGGUCUCUACGACUGCUGUGAAUACUUGGGCAGAAGUGGCAGGAGCAGCGGCAAAAAGUAAAAUGAAGCCGAAAGGC .((((...((((((..(..(-----------(....))--..)..))))))....(((((.(...(((((......)))))...).)))))))))...............(((....))) ( -31.90) >DroSec_CAF1 21913 107 + 1 AGCUGAAAGGGGGCUGAAAG-----------CAGGACG--GGUGGGGCUCUACGACUGCUGUGAAUACUUGGGCAGAAGUGGCAGGAGCAGCGGCAAAAAGUGAAAUGAAGCCGAAAGGC .((((....((((((....(-----------(......--.))..))))))....(((((.(...(((((......)))))...).)))))))))...............(((....))) ( -29.80) >DroSim_CAF1 10440 107 + 1 AGCUGAAAGGGGGCUGAAAG-----------CAGGAGG--GGUGGGGCUCUACGACUGCUGUGAAUACUUGGGCAGAAGUGGCAGGAGCAGCGGCAAAAAGUGAAAUGAAGCCGAAAGGC .((((....((((((....(-----------(......--.))..))))))....(((((.(...(((((......)))))...).)))))))))...............(((....))) ( -29.80) >DroEre_CAF1 22663 115 + 1 AGCUGAAAGGGGGCUGAAAGAGGGCUGAAAGGGGGAA-----GGGGACUCUGCAACUGCUGUGAAUACUUGGGCAGAAGUGGCAGGAGCAGUGGCAAAAAGUGAAAUGAAGCCGAAAGGC .(((.......((((.......)))).....(.(((.-----(....)))).).((((((.(...(((((......)))))...).)))))))))...............(((....))) ( -30.30) >DroYak_CAF1 12185 96 + 1 ------------------------CUGAAAGUAGGGAGUUGGGGUGGCAGUACGACUGCUGUGAAUACUCGGGCAGAAGUGGCAGGAGCGGCGGCAAAAAGUGAAAUGAAGCCGAAAGGC ------------------------...........((((....(..((((.....))))..)....))))..((....((.((....)).)).))...............(((....))) ( -25.70) >consensus AGCUGAAAGGGGGCUGAAAG___________CAGGAAG__GGUGGGGCUCUACGACUGCUGUGAAUACUUGGGCAGAAGUGGCAGGAGCAGCGGCAAAAAGUGAAAUGAAGCCGAAAGGC ....................................................((.(((((.(...(((((......)))))...).))))))).................(((....))) (-20.16 = -19.88 + -0.28)


| Location | 18,954,006 – 18,954,126 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.25 |
| Mean single sequence MFE | -32.00 |
| Consensus MFE | -26.92 |
| Energy contribution | -26.84 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18954006 120 + 22224390 GGUGGGUCUCUACGACUGCUGUGAAUACUUGGGCAGAAGUGGCAGGAGCAGCGGCAAAAAGUAAAAUGAAGCCGAAAGGCAAAUAGAGCAUAAAUUACGCAAAUCCAACACGACACUUUA (.((((((((((((.(((((.(...(((((......)))))...).))))))).................(((....)))...)))))..............))))).)........... ( -29.13) >DroSec_CAF1 21940 120 + 1 GGUGGGGCUCUACGACUGCUGUGAAUACUUGGGCAGAAGUGGCAGGAGCAGCGGCAAAAAGUGAAAUGAAGCCGAAAGGCAAAUAGAGCAUAAAUUACGCAAAUCCAACACGACACUUUA ((((..((((((((.(((((.(...(((((......)))))...).))))))).................(((....)))...))))))........((...........)).))))... ( -32.80) >DroSim_CAF1 10467 120 + 1 GGUGGGGCUCUACGACUGCUGUGAAUACUUGGGCAGAAGUGGCAGGAGCAGCGGCAAAAAGUGAAAUGAAGCCGAAAGGCAAAUAGAGCAUAAAUUACGCAAAUCCAACACGACACUUUA ((((..((((((((.(((((.(...(((((......)))))...).))))))).................(((....)))...))))))........((...........)).))))... ( -32.80) >DroEre_CAF1 22700 118 + 1 --GGGGACUCUGCAACUGCUGUGAAUACUUGGGCAGAAGUGGCAGGAGCAGUGGCAAAAAGUGAAAUGAAGCCGAAAGGCAAAUAGAGCAUAAAUUACGCAAAUCCAACACGACACUUUA --(....)..(((.((((((.(...(((((......)))))...).)))))).))).((((((.......(((....))).......((.........)).............)))))). ( -32.70) >DroYak_CAF1 12201 120 + 1 GGGGUGGCAGUACGACUGCUGUGAAUACUCGGGCAGAAGUGGCAGGAGCGGCGGCAAAAAGUGAAAUGAAGCCGAAAGGCAAAUAGAGCAUAAAUUACGCAAAUCCAACACGACACUUUA ((((((((..(((..(((((.(((....))))))))..))))).(((...(((.......(((.......(((....)))........)))......)))...))).......)))))). ( -32.58) >consensus GGUGGGGCUCUACGACUGCUGUGAAUACUUGGGCAGAAGUGGCAGGAGCAGCGGCAAAAAGUGAAAUGAAGCCGAAAGGCAAAUAGAGCAUAAAUUACGCAAAUCCAACACGACACUUUA ............((.(((((.(...(((((......)))))...).)))))))....((((((.......(((....))).......((.........)).............)))))). (-26.92 = -26.84 + -0.08)

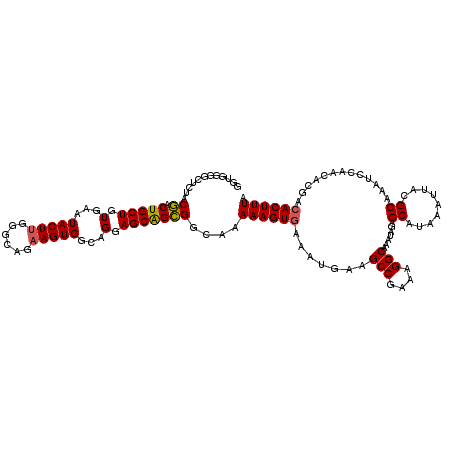
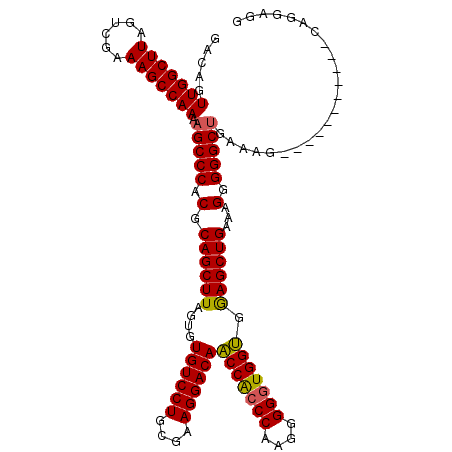
| Location | 18,954,046 – 18,954,164 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.19 |
| Mean single sequence MFE | -35.50 |
| Consensus MFE | -35.00 |
| Energy contribution | -34.72 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.72 |
| SVM RNA-class probability | 0.999942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18954046 118 + 22224390 GGCAGGAGCAGCGGCAAAAAGUAAAAUGAAGCCGAAAGGCAAAUAGAGCAUAAAUUACGCAAAUCCAACACGACACUUUACAGCGUCGACUGCGGCAGAGGCAGCAUCCUGUCCG--UGU (((((((((((((((.....(((((.((..(((....))).......((.........)).............)).)))))...)))).)))).((....))....)))))))..--... ( -35.10) >DroSec_CAF1 21980 113 + 1 GGCAGGAGCAGCGGCAAAAAGUGAAAUGAAGCCGAAAGGCAAAUAGAGCAUAAAUUACGCAAAUCCAACACGACACUUUACAGCGUCGACUGCGGCAGAGGCAGCAUCCUGUC------- (((((((((((((((..((((((.......(((....))).......((.........)).............)))))).....)))).)))).((....))....)))))))------- ( -35.70) >DroSim_CAF1 10507 114 + 1 GGCAGGAGCAGCGGCAAAAAGUGAAAUGAAGCCGAAAGGCAAAUAGAGCAUAAAUUACGCAAAUCCAACACGACACUUUACAGCGUCGACUGCGGCAGAGGCAGCAUCCUGUCC------ (((((((((((((((..((((((.......(((....))).......((.........)).............)))))).....)))).)))).((....))....))))))).------ ( -36.50) >DroEre_CAF1 22738 118 + 1 GGCAGGAGCAGUGGCAAAAAGUGAAAUGAAGCCGAAAGGCAAAUAGAGCAUAAAUUACGCAAAUCCAACACGACACUUUACAGCGUCGACUGCGGCAGAGGCAGCAUCCUGUCAG--UGU (((((((((((((((..((((((.......(((....))).......((.........)).............)))))).....))).))))).((....))....)))))))..--... ( -35.90) >DroYak_CAF1 12241 120 + 1 GGCAGGAGCGGCGGCAAAAAGUGAAAUGAAGCCGAAAGGCAAAUAGAGCAUAAAUUACGCAAAUCCAACACGACACUUUACAGCGUCGACUGCGGCAGAGGCAGCAUCCUGUUCGCAUGC (((((((((.((.((..((((((.......(((....))).......((.........)).............))))))...))((((....))))....)).)).)))))))....... ( -34.30) >consensus GGCAGGAGCAGCGGCAAAAAGUGAAAUGAAGCCGAAAGGCAAAUAGAGCAUAAAUUACGCAAAUCCAACACGACACUUUACAGCGUCGACUGCGGCAGAGGCAGCAUCCUGUCCG__UG_ (((((((((((((((..((((((.......(((....))).......((.........)).............)))))).....)))).)))).((....))....)))))))....... (-35.00 = -34.72 + -0.28)



| Location | 18,954,046 – 18,954,164 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.19 |
| Mean single sequence MFE | -32.99 |
| Consensus MFE | -31.36 |
| Energy contribution | -31.24 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.566144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18954046 118 - 22224390 ACA--CGGACAGGAUGCUGCCUCUGCCGCAGUCGACGCUGUAAAGUGUCGUGUUGGAUUUGCGUAAUUUAUGCUCUAUUUGCCUUUCGGCUUCAUUUUACUUUUUGCCGCUGCUCCUGCC ...--.((.(((((.((.((.......((((.(((((((....)))))))...((((...(((((...))))))))).)))).....(((...............))))).))))))))) ( -33.86) >DroSec_CAF1 21980 113 - 1 -------GACAGGAUGCUGCCUCUGCCGCAGUCGACGCUGUAAAGUGUCGUGUUGGAUUUGCGUAAUUUAUGCUCUAUUUGCCUUUCGGCUUCAUUUCACUUUUUGCCGCUGCUCCUGCC -------..(((((.((.......)).(((((.((((((....))))))(((.((((...(((((...))))).......(((....)))))))...)))........)))))))))).. ( -32.50) >DroSim_CAF1 10507 114 - 1 ------GGACAGGAUGCUGCCUCUGCCGCAGUCGACGCUGUAAAGUGUCGUGUUGGAUUUGCGUAAUUUAUGCUCUAUUUGCCUUUCGGCUUCAUUUCACUUUUUGCCGCUGCUCCUGCC ------((.(((((.((.......)).(((((.((((((....))))))(((.((((...(((((...))))).......(((....)))))))...)))........)))))))))))) ( -34.50) >DroEre_CAF1 22738 118 - 1 ACA--CUGACAGGAUGCUGCCUCUGCCGCAGUCGACGCUGUAAAGUGUCGUGUUGGAUUUGCGUAAUUUAUGCUCUAUUUGCCUUUCGGCUUCAUUUCACUUUUUGCCACUGCUCCUGCC ...--....(((((.((.......)).(((((.((((((....))))))(((.((((...(((((...))))).......(((....)))))))...)))........)))))))))).. ( -32.20) >DroYak_CAF1 12241 120 - 1 GCAUGCGAACAGGAUGCUGCCUCUGCCGCAGUCGACGCUGUAAAGUGUCGUGUUGGAUUUGCGUAAUUUAUGCUCUAUUUGCCUUUCGGCUUCAUUUCACUUUUUGCCGCCGCUCCUGCC (((.(((...(((......)))..((.((((..((((((....))))))(((.((((...(((((...))))).......(((....)))))))...)))...)))).)))))...))). ( -31.90) >consensus _CA__CGGACAGGAUGCUGCCUCUGCCGCAGUCGACGCUGUAAAGUGUCGUGUUGGAUUUGCGUAAUUUAUGCUCUAUUUGCCUUUCGGCUUCAUUUCACUUUUUGCCGCUGCUCCUGCC .........(((((.((.......)).(((((.((((((....))))))(((.((((...(((((...))))).......(((....)))))))...)))........)))))))))).. (-31.36 = -31.24 + -0.12)

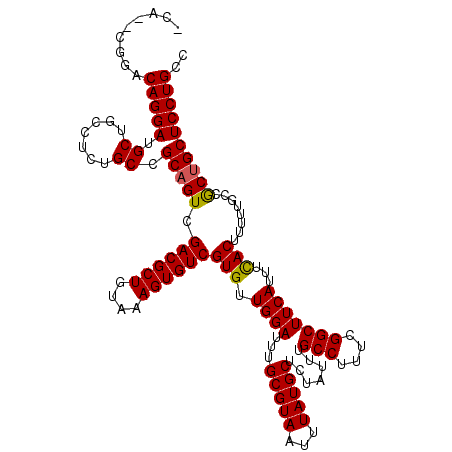

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:12 2006