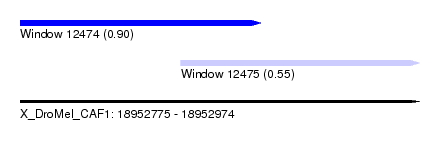
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,952,775 – 18,952,974 |
| Length | 199 |
| Max. P | 0.903704 |

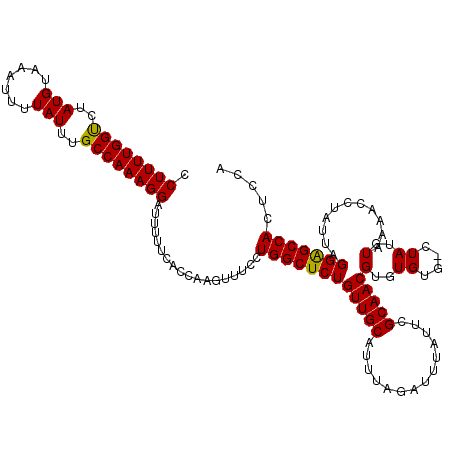
| Location | 18,952,775 – 18,952,895 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.99 |
| Mean single sequence MFE | -26.44 |
| Consensus MFE | -22.04 |
| Energy contribution | -21.72 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903704 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18952775 120 + 22224390 ACACGAAAACUCAAUAAAUUUUCAAAUAAAUUCACAUGGGCCGAGCCGAAAGAAGAGAAAUCGAAAAAACGUCCGCCUGGCCUUUUGGUCUAUGUAAAUUUUAUUUGCCAAAGGAUUUUU ....(((((.........)))))....((((((....((((((.(((....)..((....))............)).))))))((((((..(((.......)))..)))))))))))).. ( -24.40) >DroSec_CAF1 20949 119 + 1 ACACGAAAACUCAAUAAAUUUUCAAAUAAAUUCACAUGGGCCGAGCCGAAAGAAGAGAAAUCGAAAAAA-GUCCGCCUGGCCUUUUGGUCUAUGUAAAUUUUAUUUGCCAAAGGAUUUUU ...............(((((((((((((((...(((((((((....(....)..((....))...((((-(.((....)).))))))))))))))....))))))))....))))))).. ( -24.70) >DroSim_CAF1 9451 119 + 1 ACACGAAAACUCAAUAAAUUUUCAAAUAAAUUCACAUGGGCCGAGCCGAAAGAAGAGAAAUCGAAAAAA-GUCCGCCUGGCCUUUUGGUCUAUGUAAAUUUUAUUUGCCAAAGGAUUUUU ...............(((((((((((((((...(((((((((....(....)..((....))...((((-(.((....)).))))))))))))))....))))))))....))))))).. ( -24.70) >DroEre_CAF1 21689 119 + 1 ACACGAAAACUCAAUAAAUUUUCAAAUAAAUUCACAUGGGCCGAGGCACAAGAAGAGAAAUCGAAAAAA-GUCCGCCUGGCCUUUUGGCCCAUGUAAAUUUUAUUUGCCAAAGGAUUUUU ...............(((((((((((((((...((((((((((((((...((..((....))((.....-.))...)).)))))..)))))))))....))))))))....))))))).. ( -31.20) >DroYak_CAF1 11365 120 + 1 ACACGAAAACUCAAUAAAUUUUCAAAUAAAUUCACAUGGGCCGAGGCGAAAGCAGAGAAAUCGAAAAAACGUCCCCCUGGCCUUUUGGUCUAUGUAAAUUUUAUUUGCCAAAGGAUUUUU ...............(((((((((((((((...((((((((((((((....)).((....)).....................))))))))))))....))))))))....))))))).. ( -27.20) >consensus ACACGAAAACUCAAUAAAUUUUCAAAUAAAUUCACAUGGGCCGAGCCGAAAGAAGAGAAAUCGAAAAAA_GUCCGCCUGGCCUUUUGGUCUAUGUAAAUUUUAUUUGCCAAAGGAUUUUU ...............(((((((((((((((...((((((((((((.(....)..................(((.....)))..))))))))))))....))))))))....))))))).. (-22.04 = -21.72 + -0.32)

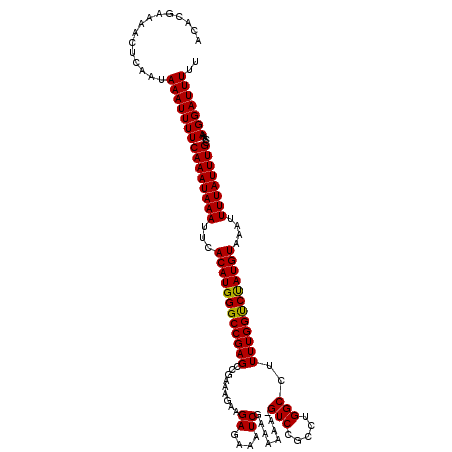
| Location | 18,952,855 – 18,952,974 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.49 |
| Mean single sequence MFE | -26.44 |
| Consensus MFE | -24.26 |
| Energy contribution | -23.86 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18952855 119 + 22224390 CCUUUUGGUCUAUGUAAAUUUUAUUUGCCAAAGGAUUUUUCACCAAGUUUCCUGGCUCUGUUGCAUUUAGAUUUAUUCGCAACGUGUGUG-CUAUGUAAAACCUAUUAGGAGCCACUCCA .((((((((..(((.......)))..))))))))..................((((((((((((..............)))))((...((-(...)))..))......)))))))..... ( -25.44) >DroSec_CAF1 21028 119 + 1 CCUUUUGGUCUAUGUAAAUUUUAUUUGCCAAAGGAUUUUUCACCAAGUUUCCUGGCUCUGUUGCAUUUAGAUUUAUUCGCAACGUGUGUG-CUAUGUAAAACCUAUUAGGAGCCACUCCA .((((((((..(((.......)))..))))))))..................((((((((((((..............)))))((...((-(...)))..))......)))))))..... ( -25.44) >DroSim_CAF1 9530 119 + 1 CCUUUUGGUCUAUGUAAAUUUUAUUUGCCAAAGGAUUUUUCACCAAGUUUCCUGGCUCUGUUGCAUUUAGAUUUAUUCGCAACGUGUGUG-CUAUGUAAAACCUAUUAGGAGCCACUCCA .((((((((..(((.......)))..))))))))..................((((((((((((..............)))))((...((-(...)))..))......)))))))..... ( -25.44) >DroEre_CAF1 21768 119 + 1 CCUUUUGGCCCAUGUAAAUUUUAUUUGCCAAAGGAUUUUUCACCAAGUUUCCUGGCUCUGUUGCAUUUAGAUUUAUUCGCAACGUGUGUG-CUAUGUAAAACCUAUUAGGGGCCACUCCA .....((((((..(((..((((((..((...((((..(((....)))..)))).((.(((((((..............)))))).).)))-)...))))))..)))...))))))..... ( -28.14) >DroYak_CAF1 11445 120 + 1 CCUUUUGGUCUAUGUAAAUUUUAUUUGCCAAAGGAUUUUUCACCAAGUUUCCUGGCUCUGUUGCAUUUAGAUUUAUUCGCAACGUGUGUGGCUAUGUAAAACCUAUUAGGGGCCACUCCA .....((((((..(((..((((((..(((..((((..(((....)))..)))).((.(((((((..............)))))).).)))))...))))))..)))...))))))..... ( -27.74) >consensus CCUUUUGGUCUAUGUAAAUUUUAUUUGCCAAAGGAUUUUUCACCAAGUUUCCUGGCUCUGUUGCAUUUAGAUUUAUUCGCAACGUGUGUG_CUAUGUAAAACCUAUUAGGAGCCACUCCA .((((((((..(((.......)))..))))))))..................((((((((((((..............)))))(..((....))..)...........)))))))..... (-24.26 = -23.86 + -0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:03 2006