
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,952,547 – 18,952,690 |
| Length | 143 |
| Max. P | 0.998558 |

| Location | 18,952,547 – 18,952,655 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -28.62 |
| Consensus MFE | -25.78 |
| Energy contribution | -25.86 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.14 |
| SVM RNA-class probability | 0.998558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
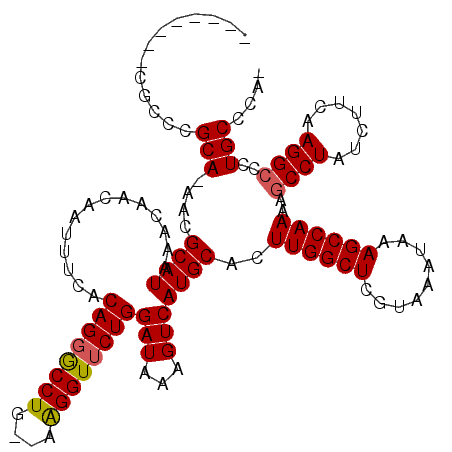
>X_DroMel_CAF1 18952547 108 + 22224390 --------CGCCCGCA-AACGCAUAAACAACAAUUUCACAGGGCCUG--AAGGUUCUGGAUAAAGUCAUGCACUUGGCUCGUAAAUAAAGCCAAAAGCCUAUCUUCAAGGCCCUGCCCA- --------.....((.-...))................(((((((((--(((((...((.....(.....)..((((((.........))))))...)).)))))).))))))))....- ( -30.90) >DroSec_CAF1 20711 108 + 1 --------CGCCCGCA-AACGCAUAAACAACAAUUUCACAGGGCCUG--AAGGUUCUGGAUAAAGUCAUGCACUUGGCUCGUAAAUAAAGCCAAAAGCCUAUCUUCAAGGCCCUGCCCA- --------.....((.-...))................(((((((((--(((((...((.....(.....)..((((((.........))))))...)).)))))).))))))))....- ( -30.90) >DroSim_CAF1 9223 108 + 1 --------CGCCCGCA-AACGCAUAAACAACAAUUUCACAGGGCCUG--AAGGUUCUGGAUAAAGUCAUGCACUUGGCUCGUAAAUAAAGCCAAAAGCCUAUCUUCAAGGCCCUGCCCA- --------.....((.-...))................(((((((((--(((((...((.....(.....)..((((((.........))))))...)).)))))).))))))))....- ( -30.90) >DroEre_CAF1 21437 108 + 1 --------CGCCCGCA-AACGCAUAAACAACAAUUUCACAGCGCCUG--AAGGUUCUGGAUAAAGUCAUGCACUUGGCUCGUAAAUAAAGCCAAAAGCCUAUCUUCAAGGCCCUGCCCA- --------.....((.-...))................(((.(((((--(((((...((.....(.....)..((((((.........))))))...)).)))))).)))).)))....- ( -23.80) >DroYak_CAF1 11116 120 + 1 GCCGCAAACGCCCGCAAAACGCAUAAACAACAAUUUCACAGGACCUGGCAGGGUUCUGGAUAAAGUCAUGCACUUGGCUCGUAAAUAAAGCCAAAAGCCUAUCUUCAAGGACCUGCCCAC .((((....))..((.....))..................))....((((((.((((.((...((....((..((((((.........))))))..))....)))).))))))))))... ( -26.60) >consensus ________CGCCCGCA_AACGCAUAAACAACAAUUUCACAGGGCCUG__AAGGUUCUGGAUAAAGUCAUGCACUUGGCUCGUAAAUAAAGCCAAAAGCCUAUCUUCAAGGCCCUGCCCA_ .............(((....((((..............((((((((....))))))))(((...)))))))..((((((.........))))))..((((.......))))..))).... (-25.78 = -25.86 + 0.08)

| Location | 18,952,578 – 18,952,690 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.62 |
| Mean single sequence MFE | -34.80 |
| Consensus MFE | -28.48 |
| Energy contribution | -28.92 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
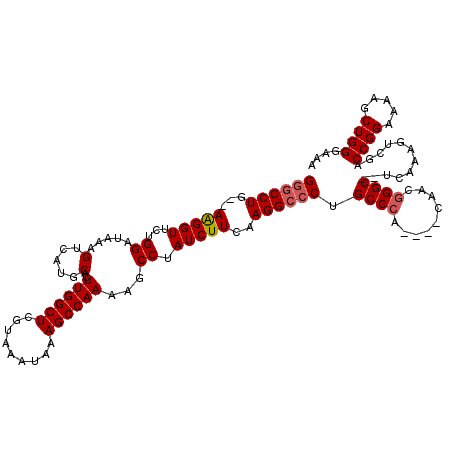
>X_DroMel_CAF1 18952578 112 + 22224390 GGGCCUG--AAGGUUCUGGAUAAAGUCAUGCACUUGGCUCGUAAAUAAAGCCAAAAGCCUAUCUUCAAGGCCCUGCCCA----CCACGGGC--UCAAAGUCGACCGGAAAAGCUGGGAAA (((((((--(((((...((.....(.....)..((((((.........))))))...)).)))))).)))))).((((.----....))))--..........((((.....)))).... ( -37.10) >DroSec_CAF1 20742 112 + 1 GGGCCUG--AAGGUUCUGGAUAAAGUCAUGCACUUGGCUCGUAAAUAAAGCCAAAAGCCUAUCUUCAAGGCCCUGCCCA----CAACGGGC--UCAAAGUCGACCGGAAGAGCUGGGAAA (((((((--(((((...((.....(.....)..((((((.........))))))...)).)))))).)))))).((((.----....))))--..........((((.....)))).... ( -37.10) >DroSim_CAF1 9254 112 + 1 GGGCCUG--AAGGUUCUGGAUAAAGUCAUGCACUUGGCUCGUAAAUAAAGCCAAAAGCCUAUCUUCAAGGCCCUGCCCA----CAACGGGC--UCAAAGUCGACCGGAAGAGCUGGGAAA (((((((--(((((...((.....(.....)..((((((.........))))))...)).)))))).)))))).((((.----....))))--..........((((.....)))).... ( -37.10) >DroEre_CAF1 21468 112 + 1 GCGCCUG--AAGGUUCUGGAUAAAGUCAUGCACUUGGCUCGUAAAUAAAGCCAAAAGCCUAUCUUCAAGGCCCUGCCCA----CAACGGGC--UCAAAGUCGACCGGAAAAGCUGGGAAA ..(((((--(((((...((.....(.....)..((((((.........))))))...)).)))))).))))...((((.----....))))--..........((((.....)))).... ( -30.40) >DroYak_CAF1 11156 120 + 1 GGACCUGGCAGGGUUCUGGAUAAAGUCAUGCACUUGGCUCGUAAAUAAAGCCAAAAGCCUAUCUUCAAGGACCUGCCCACACACAACAGGCUCUCAAAGUCGACCGGAAAAGCUGGGAAA ...((..((....((((((..........((..((((((.........))))))..))....(((..((..((((...........))))..))..)))....))))))..))..))... ( -32.30) >consensus GGGCCUG__AAGGUUCUGGAUAAAGUCAUGCACUUGGCUCGUAAAUAAAGCCAAAAGCCUAUCUUCAAGGCCCUGCCCA____CAACGGGC__UCAAAGUCGACCGGAAAAGCUGGGAAA ((((((...(((((...((.....(.....)..((((((.........))))))...)).)))))..)))))).((((.........))))............((((.....)))).... (-28.48 = -28.92 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:01 2006