| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,934,639 – 18,934,732 |
| Length | 93 |
| Max. P | 0.960562 |

| Location | 18,934,639 – 18,934,732 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 78.82 |
| Mean single sequence MFE | -25.85 |
| Consensus MFE | -18.75 |
| Energy contribution | -18.75 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18934639 93 + 22224390 GCGCAUAUAUUUCGCAGACUUUCGCCA--CCCCCUCCCCGAUUUCAGUUGCUGCUUUUGACAGUUUUGUGUACUGUCAAUUUAAGGCAGCGGUGU- (((.(.....).))).......((((.--..........(....)....(((((((((((((((.......)))))))....)))))))))))).- ( -25.40) >DroPse_CAF1 210391 82 + 1 GCGCAUAUAUUUCACCGAGUUGC-------------CUC-GUUCUUGUGGCUGCUUUUGACAGUUUUGUGUACUGUCAAUUUGAGGCAGAAGUGCU ((((.......((((((((....-------------)))-).....))))((((((((((((((.......)))))))....)))))))..)))). ( -26.40) >DroYak_CAF1 160913 95 + 1 GCGCAUAUAUUUCGCAGACUUUCGCCAAGCCCCAACCCCGAUUUCAGUUGCUGCUUUUGACAGUUUUGUGUACUGUCAAUUUAAGGCAGCAGUGU- ((((.........((........)).......................((((((((((((((((.......)))))))....)))))))))))))- ( -24.20) >DroPer_CAF1 210772 82 + 1 GCGCAUAUAUUUCACCGAGCUGC-------------CUC-GUUCUUGUGGCUGCUUUUGACAGUUUUGUGUACUGUCAAUUUGAGGCAGAAGUGCU ((((.......((((.((((...-------------...-))))..))))((((((((((((((.......)))))))....)))))))..)))). ( -27.40) >consensus GCGCAUAUAUUUCACAGACUUGC_____________CCC_AUUCCAGUGGCUGCUUUUGACAGUUUUGUGUACUGUCAAUUUAAGGCAGAAGUGC_ ((((............(.....)..........................(((((((((((((((.......)))))))....)))))))).)))). (-18.75 = -18.75 + 0.00)

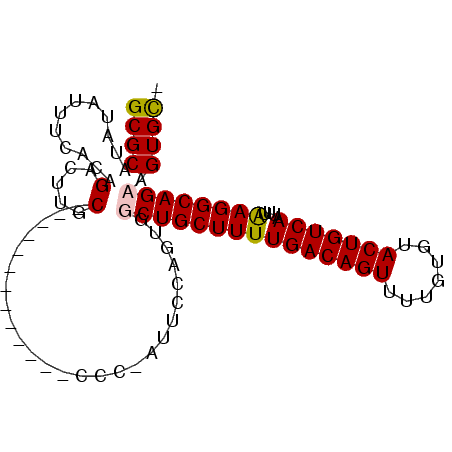

| Location | 18,934,639 – 18,934,732 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 78.82 |
| Mean single sequence MFE | -25.23 |
| Consensus MFE | -13.07 |
| Energy contribution | -13.58 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18934639 93 - 22224390 -ACACCGCUGCCUUAAAUUGACAGUACACAAAACUGUCAAAAGCAGCAACUGAAAUCGGGGAGGGGG--UGGCGAAAGUCUGCGAAAUAUAUGCGC -.(((((((((.((...((((((((.......)))))))))))))))..(((....)))......))--))((....))..(((.........))) ( -27.70) >DroPse_CAF1 210391 82 - 1 AGCACUUCUGCCUCAAAUUGACAGUACACAAAACUGUCAAAAGCAGCCACAAGAAC-GAG-------------GCAACUCGGUGAAAUAUAUGCGC .(((...((((......((((((((.......))))))))..))))...((....(-(((-------------....)))).)).......))).. ( -22.90) >DroYak_CAF1 160913 95 - 1 -ACACUGCUGCCUUAAAUUGACAGUACACAAAACUGUCAAAAGCAGCAACUGAAAUCGGGGUUGGGGCUUGGCGAAAGUCUGCGAAAUAUAUGCGC -....((((((.((...((((((((.......)))))))))))))))).(((....))).......((..(((....))).))............. ( -26.40) >DroPer_CAF1 210772 82 - 1 AGCACUUCUGCCUCAAAUUGACAGUACACAAAACUGUCAAAAGCAGCCACAAGAAC-GAG-------------GCAGCUCGGUGAAAUAUAUGCGC ..((((.(((((((...((((((((.......))))))))..(......)......-)))-------------))))...))))............ ( -23.90) >consensus _ACACUGCUGCCUCAAAUUGACAGUACACAAAACUGUCAAAAGCAGCAACAAAAAC_GAG_____________GAAACUCGGCGAAAUAUAUGCGC ......(((((......((((((((.......))))))))..)))))................................................. (-13.07 = -13.58 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:54 2006