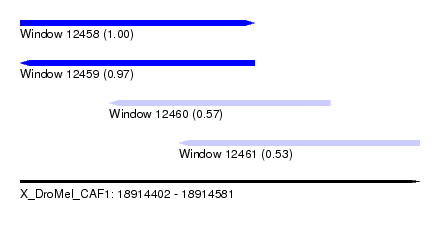
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,914,402 – 18,914,581 |
| Length | 179 |
| Max. P | 0.996424 |

| Location | 18,914,402 – 18,914,507 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 61.01 |
| Mean single sequence MFE | -29.56 |
| Consensus MFE | -14.27 |
| Energy contribution | -16.98 |
| Covariance contribution | 2.71 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.48 |
| SVM decision value | 2.70 |
| SVM RNA-class probability | 0.996424 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18914402 105 + 22224390 UUAAGCGUGAUCAUUUCUAUUAAAGUAAAGUACCUGGUUAUUAAAAUGCCCAGGUA---------CACCGCCCAUUUGAUAUCCACCACUAGA-----UGAUGGUGCGAUCGCUACCUG- ......((((((.................((((((((.(((....))).)))))))---------)...(((((((...((........))..-----.))))).))))))))......- ( -29.90) >DroSec_CAF1 147818 105 + 1 UCAAGCGUGAUCAUUUCUAUUAAAGUAAAGUACCUGGUUAUUAAAAUGCCCAGGUA---------CACCGACCAUUUGAUACCCACCACUAGA-----UGGUGGUGCGAUCGCUACCUU- ......((((((.....(((((((((...((((((((.(((....))).)))))))---------)....))..)))))))..(((((((...-----.))))))).))))))......- ( -33.70) >DroSim_CAF1 121280 99 + 1 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN---------NNCCCGCUCUUUGUU-CUCACC--CUGA-----UGGGGG--CGA-CGCUACCUU- ........................................................---------....(((((((...(-(.....--..))-----.)))))--)).-.........- ( -9.20) >DroEre_CAF1 141974 104 + 1 UGAAGCGUGACCACUUCUAUCAAAGUAAAGUACCUGGUUAUUAAAGUGCCCAGGUA---------CACCACCCAUUUAAUGGCCACCACUAGCG------GUGGUGCGAUCGCUACCUG- ...((((((...((((......))))...((((((((.(((....))).)))))))---------)..)))..........((((((......)------)))))......))).....- ( -30.90) >DroYak_CAF1 142927 120 + 1 UUCAGCGUGAUCAUUUCUAUUAAAGUAAAGUACCUGGUUAUUAAGAUGCCCAGGUACGAGUACUACACCACCCAUUCAAUCCCCACCACUGGUGGUGCUGGUGGUGCGAUCGCUACCUGG ..(((.((((((...........((((..((((((((.(((....))).))))))))...)))).(((((((....((....(((((...)))))))..))))))).))))))...))). ( -44.10) >consensus UUAAGCGUGAUCAUUUCUAUUAAAGUAAAGUACCUGGUUAUUAAAAUGCCCAGGUA_________CACCACCCAUUUGAUACCCACCACUAGA_____UGGUGGUGCGAUCGCUACCUG_ ......((((((..................(((((((.(((....))).)))))))...........................(((((((.........))))))).))))))....... (-14.27 = -16.98 + 2.71)

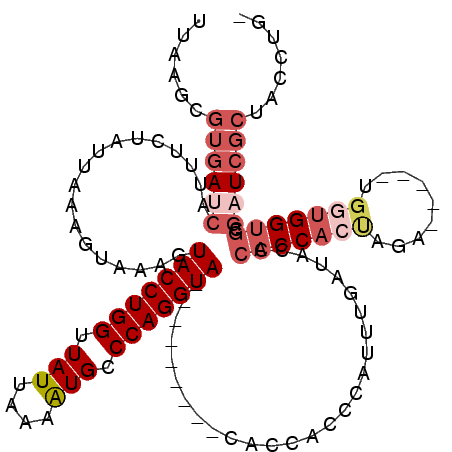
| Location | 18,914,402 – 18,914,507 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 61.01 |
| Mean single sequence MFE | -30.96 |
| Consensus MFE | -12.76 |
| Energy contribution | -14.32 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.41 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18914402 105 - 22224390 -CAGGUAGCGAUCGCACCAUCA-----UCUAGUGGUGGAUAUCAAAUGGGCGGUG---------UACCUGGGCAUUUUAAUAACCAGGUACUUUACUUUAAUAGAAAUGAUCACGCUUAA -..((((((....)).((((((-----(...))))))).))))...((((((..(---------(((((((............))))))))..........((....))....)))))). ( -30.70) >DroSec_CAF1 147818 105 - 1 -AAGGUAGCGAUCGCACCACCA-----UCUAGUGGUGGGUAUCAAAUGGUCGGUG---------UACCUGGGCAUUUUAAUAACCAGGUACUUUACUUUAAUAGAAAUGAUCACGCUUGA -.....(((((((.((((((..-----....)))))).).)))...((((((..(---------(((((((............))))))))....((.....))...)))))).)))... ( -33.40) >DroSim_CAF1 121280 99 - 1 -AAGGUAGCG-UCG--CCCCCA-----UCAG--GGUGAG-AACAAAGAGCGGGNN---------NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN -.........-(((--(((...-----...)--))))).-...............---------........................................................ ( -9.00) >DroEre_CAF1 141974 104 - 1 -CAGGUAGCGAUCGCACCAC------CGCUAGUGGUGGCCAUUAAAUGGGUGGUG---------UACCUGGGCACUUUAAUAACCAGGUACUUUACUUUGAUAGAAGUGGUCACGCUUCA -.((((((..(((((.((((------((....))))))((((...)))))))))(---------(((((((............)))))))).)))))).....((((((....)))))). ( -39.10) >DroYak_CAF1 142927 120 - 1 CCAGGUAGCGAUCGCACCACCAGCACCACCAGUGGUGGGGAUUGAAUGGGUGGUGUAGUACUCGUACCUGGGCAUCUUAAUAACCAGGUACUUUACUUUAAUAGAAAUGAUCACGCUGAA .(((...(.((((((((((((..(((((((...)))))....))....))))))))((((...((((((((............))))))))..))))...........)))).).))).. ( -42.60) >consensus _CAGGUAGCGAUCGCACCACCA_____UCUAGUGGUGGGUAUCAAAUGGGCGGUG_________UACCUGGGCAUUUUAAUAACCAGGUACUUUACUUUAAUAGAAAUGAUCACGCUUAA .........((((.((((((...........))))))...........................(((((((............)))))))..................))))........ (-12.76 = -14.32 + 1.56)

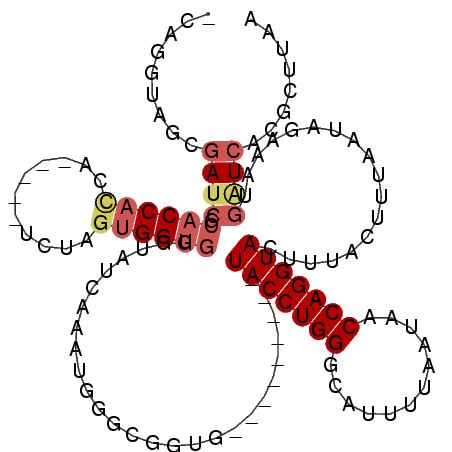

| Location | 18,914,442 – 18,914,541 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.34 |
| Mean single sequence MFE | -31.60 |
| Consensus MFE | -19.06 |
| Energy contribution | -19.88 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18914442 99 - 22224390 UAAACAAUAGCUGCCAGC------AACAUGAUCCAAUAAA-CAGGUAGCGAUCGCACCAUCA-----UCUAGUGGUGGAUAUCAAAUGGGCGGUG---------UACCUGGGCAUUUUAA ...........((((...------...((......))...-((((((.(.(((((.((((((-----(...)))))))...((....))))))))---------))))))))))...... ( -24.50) >DroSec_CAF1 147858 99 - 1 UAAAAAAUAGCUGCCGGC------AGCAUGAUCCAAUAAA-AAGGUAGCGAUCGCACCACCA-----UCUAGUGGUGGGUAUCAAAUGGUCGGUG---------UACCUGGGCAUUUUAA .........((((((...------................-..))))))((((.((((((..-----....)))))).).)))(((((.((((..---------...)))).)))))... ( -28.70) >DroEre_CAF1 142014 98 - 1 UAAACAAUAGCUGCCAGC------AGCAUGAUCCAAUAAA-CAGGUAGCGAUCGCACCAC------CGCUAGUGGUGGCCAUUAAAUGGGUGGUG---------UACCUGGGCACUUUAA .........((((....)------))).............-((((((.(.(((((.((((------((....))))))((((...))))))))))---------)))))).......... ( -30.60) >DroYak_CAF1 142967 120 - 1 UAAACAAUAGCUGCCAGCAGCAGCAGCAUGAUCCAAUAAACCAGGUAGCGAUCGCACCACCAGCACCACCAGUGGUGGGGAUUGAAUGGGUGGUGUAGUACUCGUACCUGGGCAUCUUAA .........(((((.....)))))................(((((((.(((.(((((((((..(((((((...)))))....))....)))))))).)...))))))))))......... ( -42.60) >consensus UAAACAAUAGCUGCCAGC______AGCAUGAUCCAAUAAA_CAGGUAGCGAUCGCACCACCA_____UCUAGUGGUGGGUAUCAAAUGGGCGGUG_________UACCUGGGCAUUUUAA ...........((((.((.......))..............((((((((....)).((((((..........))))))..........................))))))))))...... (-19.06 = -19.88 + 0.81)



| Location | 18,914,473 – 18,914,581 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.26 |
| Mean single sequence MFE | -24.44 |
| Consensus MFE | -14.72 |
| Energy contribution | -15.80 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18914473 108 - 22224390 UAUGCAAUUCAUCGAAAUCGACACAAAGUUGCAAAAAUAAUAAACAAUAGCUGCCAGC------AACAUGAUCCAAUAAA-CAGGUAGCGAUCGCACCAUCA-----UCUAGUGGUGGAU ..(((((((..(((....))).....)))))))................((((((...------................-..)))))).(((.((((((..-----....))))))))) ( -24.50) >DroSec_CAF1 147889 108 - 1 UAUGCAAUUCAUCCAAAUCGACACAAAGUUGCAAAAAUAAUAAAAAAUAGCUGCCGGC------AGCAUGAUCCAAUAAA-AAGGUAGCGAUCGCACCACCA-----UCUAGUGGUGGGU ..(((...((........((((.....))))..................((((((...------................-..))))))))..)))((((((-----(...))))))).. ( -23.70) >DroSim_CAF1 121351 91 - 1 U-UGC-AUUCAUCCA-AUCGACACAAAGUUGCAAAA-UAA--AAACAAAGCUGC-GGC------AGC-UGAUCA---AAA-AAGGUAGCG-UCG--CCCCCA-----UCAG--GGUGAG- .-...-..((((((.-..((((.....)))).....-...--..........((-(((------.((-(.(((.---...-..)))))))-)))--).....-----...)--))))).- ( -21.00) >DroEre_CAF1 142045 107 - 1 UAUGCAAUUCAUCCGAAUCGACACAAAGUUGCAAAAAUAAUAAACAAUAGCUGCCAGC------AGCAUGAUCCAAUAAA-CAGGUAGCGAUCGCACCAC------CGCUAGUGGUGGCC ..(((.((((....)))).........(((((.................((((....)------))).....((......-..))..))))).)))((((------((....)))))).. ( -23.40) >DroYak_CAF1 143007 120 - 1 UAUGCAAUUCAUCCGAAUCGACACAAAGUUGCAAAAAUAAUAAACAAUAGCUGCCAGCAGCAGCAGCAUGAUCCAAUAAACCAGGUAGCGAUCGCACCACCAGCACCACCAGUGGUGGGG (((((.............((((.....))))..................(((((.....))))).)))))..........((.(((.((....))))).....(((((....))))))). ( -29.60) >consensus UAUGCAAUUCAUCCAAAUCGACACAAAGUUGCAAAAAUAAUAAACAAUAGCUGCCAGC______AGCAUGAUCCAAUAAA_CAGGUAGCGAUCGCACCACCA_____UCUAGUGGUGGGU ..(((((((.................)))))))................((((((............................)))))).....((((((...........))))))... (-14.72 = -15.80 + 1.08)

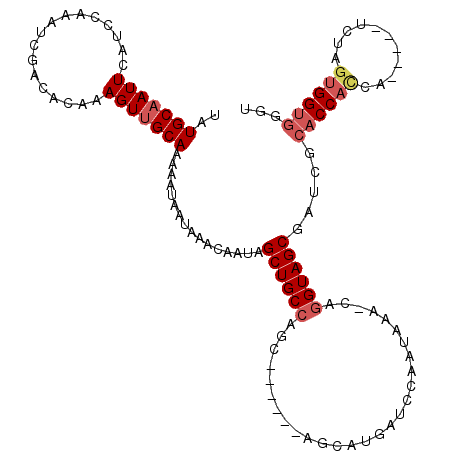

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:48 2006