
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,885,276 – 18,885,370 |
| Length | 94 |
| Max. P | 0.550157 |

| Location | 18,885,276 – 18,885,370 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.93 |
| Mean single sequence MFE | -36.02 |
| Consensus MFE | -11.97 |
| Energy contribution | -13.00 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.33 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18885276 94 + 22224390 AAGUUUUGGCCCACUGAAUGGGUU-CAAAAAGUGCCAUAAAACACC---CGCUAAACAGAA----GUUUGCGU---------GG-GGCGG---GGGC--G---GCGGCCAAAGUUGGCUU ...((((((((((.....)))).)-)))))...(((........((---((((((((....----))))).))---------))-)((..---..))--)---))(((((....))))). ( -32.60) >DroPse_CAF1 166795 119 + 1 AAGUUUU-GCCCAGUGAAUGGGUUUUAAGAAGUGCCAUAAAACACUGCGAACGAAGCAGAAAGAGCUGUGCAUCCCAACCGAGGGGGCCAGUGCGGCAGGGAGGCGACAAAAGUUGGCUU .......-(((((.....))))).........((((........((((.......))))...(.((((.((.((((......)))))))))).)))))..(((.((((....)))).))) ( -37.80) >DroSec_CAF1 121293 98 + 1 AAGUUUUGGCCCACUGAAUGGGUU-CAAAAAGAGCCAUAAAACACC---CGCUAAGCAGAA----GUUUGCAU---------GGUGGCGG---GGGC--GGGGGCGGCCAAAGUUGGCUU ..(((((.((((.(((....((((-(.....))))).........(---(((((.((((..----..)))).)---------))))))))---))))--.)))))(((((....))))). ( -39.20) >DroEre_CAF1 120121 91 + 1 AAGUUUUGGCCCACUGAAUGGGCU-CGCAAAGAGCCAUAAAACAGC---CGCUAAGCAGAA----CUUUCCGC---------GGG----------GC--GCGGACGGCCAAAGUUGGCUU ...((((((((..(((....((((-(.....)))))..........---((((..((.((.----...)).))---------..)----------))--))))..))))))))....... ( -31.00) >DroYak_CAF1 118661 97 + 1 AAGUUGUGGCCCACUGAAUGGGUU-CAAAAAGAGCCAUAAAACACC---CGCUAAGUGGAA----CUUUCCGC---------GGGGGCG----UGGC--GGGGGCGGCCAAAGUUGGCUU ........((((.(((....((((-(.....)))))........((---((((..(((((.----...)))))---------...))))----.)))--))))))(((((....))))). ( -38.40) >DroPer_CAF1 167512 119 + 1 AAGUUUU-GCCCAGUGAAUGGGUUUUAAGAAGUGCCAUAAAACACUGCGAACGAAGCAGAAAGAACUGUGCAUCCCAGCCGAGGGCGGCAGUGCGGCAGGGAGGCGACAAAAGUUGGCUU .......-(((((.....)))))..........(((........((((.......))))....((((.(((.((((.((((...((....)).)))).)))).))).....))))))).. ( -37.10) >consensus AAGUUUUGGCCCACUGAAUGGGUU_CAAAAAGAGCCAUAAAACACC___CGCUAAGCAGAA____CUUUGCAU_________GGGGGCG____CGGC__GGGGGCGGCCAAAGUUGGCUU ...((((((((((.....)))))..)))))...((((..................(((((......)))))........................((......)).........)))).. (-11.97 = -13.00 + 1.03)

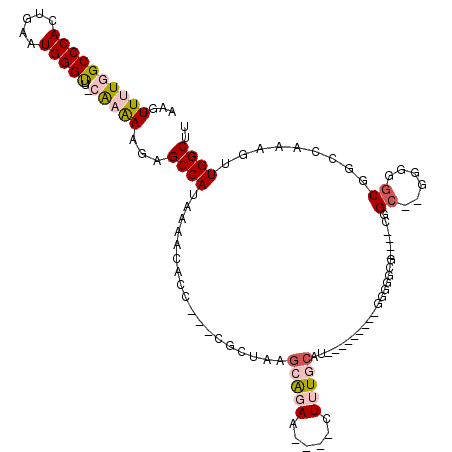

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:41 2006