
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,864,879 – 18,864,979 |
| Length | 100 |
| Max. P | 0.997020 |

| Location | 18,864,879 – 18,864,979 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 76.92 |
| Mean single sequence MFE | -35.04 |
| Consensus MFE | -11.08 |
| Energy contribution | -13.60 |
| Covariance contribution | 2.52 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.32 |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.997020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
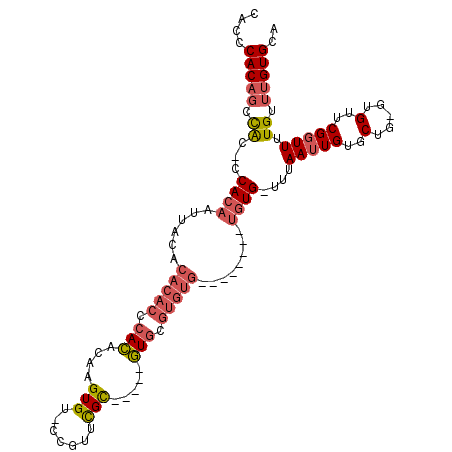
>X_DroMel_CAF1 18864879 100 + 22224390 UGCACAAACAAAACCGAACAC-CAGCACAAUUAAA-CACACAC----ACACACGCAC-----GCGAACGG-ACACUUGUGUGGGUGUGUGUAAUUGUGG-GUGGCUGUGGGUG ..................(((-(..((((.(((..-((((.((----((((((.(((-----((((....-....))))))).))))))))...)))).-.))).)))))))) ( -37.50) >DroSec_CAF1 101490 96 + 1 UGCACAAACAAAACCGAACAC-CAGCACAAUUAAA-CACA--------CACACGCAC-----GCGAACGG-ACACUUGUGUGGGUGUGUGUAAUUGUGG-GUGGCUGUGGGUG ............(((.(.(((-(..(((((((...-.(((--------(((((.(((-----((((....-....))))))).))))))))))))))))-)))....).))). ( -37.30) >DroSim_CAF1 96575 96 + 1 UGCACAAACAAAACCGAACAC-CAGCACAAUUAAA-CACA--------CACACGCAC-----GCGAACGG-ACACUUGUGUGGGUGUGUGUAAUUGUGG-GUGGCUGUGGGUG ............(((.(.(((-(..(((((((...-.(((--------(((((.(((-----((((....-....))))))).))))))))))))))))-)))....).))). ( -37.30) >DroEre_CAF1 102242 96 + 1 UGCACAAACAAAACCGAACAC-CAGCACAAUUAAA-CACA--------CACACGCAC-----GCGAACGG-ACACUUCUGUGAGUGUGUGUAAUUGUGG-GCGGCGGUGGGCG ..............((..(((-(.((.........-((((--------...((((((-----((..((((-(....)))))..))))))))...)))).-...))))))..)) ( -31.12) >DroPer_CAF1 143201 105 + 1 UACACAAACAAAACCGAACCCUCGGCCCAUUUAAGGCAUCCAUGCACACACCCAGAUGCAACACAGGUGGGGCACGAGUAUGC--------GAGUGUGGGGUAUGUGUGAGUG ((((((.((....((((....))))(((.......(((....)))((((.((((..((.....))..))))(((......)))--------..))))))))).)))))).... ( -32.00) >consensus UGCACAAACAAAACCGAACAC_CAGCACAAUUAAA_CACA________CACACGCAC_____GCGAACGG_ACACUUGUGUGGGUGUGUGUAAUUGUGG_GUGGCUGUGGGUG .............(((..(((((..((((((((...............(((((((........(....)...(((....))).))))))))))))))))))))....)))... (-11.08 = -13.60 + 2.52)



| Location | 18,864,879 – 18,864,979 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 76.92 |
| Mean single sequence MFE | -33.32 |
| Consensus MFE | -10.38 |
| Energy contribution | -11.62 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.31 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18864879 100 - 22224390 CACCCACAGCCAC-CCACAAUUACACACACCCACACAAGUGU-CCGUUCGC-----GUGCGUGUGU----GUGUGUG-UUUAAUUGUGCUG-GUGUUCGGUUUUGUUUGUGCA .(((....((((.-.((((((((.((((((.((((((...(.-.((....)-----)..).)))))----).)))))-).)))))))).))-))....)))............ ( -36.60) >DroSec_CAF1 101490 96 - 1 CACCCACAGCCAC-CCACAAUUACACACACCCACACAAGUGU-CCGUUCGC-----GUGCGUGUG--------UGUG-UUUAAUUGUGCUG-GUGUUCGGUUUUGUUUGUGCA .(((....((((.-.((((((((.((((((.(((.((.(((.-.....)))-----.)).))).)--------))))-).)))))))).))-))....)))............ ( -33.00) >DroSim_CAF1 96575 96 - 1 CACCCACAGCCAC-CCACAAUUACACACACCCACACAAGUGU-CCGUUCGC-----GUGCGUGUG--------UGUG-UUUAAUUGUGCUG-GUGUUCGGUUUUGUUUGUGCA .(((....((((.-.((((((((.((((((.(((.((.(((.-.....)))-----.)).))).)--------))))-).)))))))).))-))....)))............ ( -33.00) >DroEre_CAF1 102242 96 - 1 CGCCCACCGCCGC-CCACAAUUACACACACUCACAGAAGUGU-CCGUUCGC-----GUGCGUGUG--------UGUG-UUUAAUUGUGCUG-GUGUUCGGUUUUGUUUGUGCA .....((((.(((-(((((((((.((((((.(((....(((.-.....)))-----....))).)--------))))-).))))))))..)-)))..))))............ ( -33.10) >DroPer_CAF1 143201 105 - 1 CACUCACACAUACCCCACACUC--------GCAUACUCGUGCCCCACCUGUGUUGCAUCUGGGUGUGUGCAUGGAUGCCUUAAAUGGGCCGAGGGUUCGGUUUUGUUUGUGUA (((..(((...(((..((.(((--------(.....(((((((.(((((...........))))).).))))))..((((.....)))))))).))..)))..)))..))).. ( -30.90) >consensus CACCCACAGCCAC_CCACAAUUACACACACCCACACAAGUGU_CCGUUCGC_____GUGCGUGUG________UGUG_UUUAAUUGUGCUG_GUGUUCGGUUUUGUUUGUGCA ....(((((.((...((((......(((((.(((....(((.......))).....))).)))))........))))....(((((..(.....)..))))).)).))))).. (-10.38 = -11.62 + 1.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:33 2006