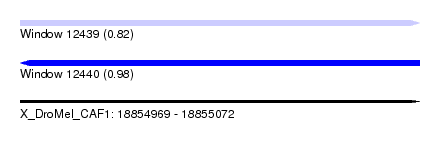
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,854,969 – 18,855,072 |
| Length | 103 |
| Max. P | 0.977293 |

| Location | 18,854,969 – 18,855,072 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 92.04 |
| Mean single sequence MFE | -31.47 |
| Consensus MFE | -28.50 |
| Energy contribution | -28.88 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18854969 103 + 22224390 AAGGCGGACGUUGGAAUGGCACUUGGUUUUUCGUUGUUCUU-GCCAUUAUUAUGGCCAUUGUUGUUGCGCCGUGCCGCUUCACACCUUUAUUUGUAUUGGUAU------U .(((((((((.((.((..(((...((....)).........-(((((....)))))...)))..)).)).))).))))))...(((..((....))..)))..------. ( -26.60) >DroSec_CAF1 91534 104 + 1 AAGGCGGGCGACGGAAUGGCACUUGGUUUUUCGUUGUUCCUGGCCAUUAUUAUGUCCAUUGUUGUUGCGCCGUGCCGCUUCACACCUUUAUUUGUAUUGGUAU------U ..((((((((((((((..((............))..))))(((.(((....))).)))..)))))).))))((((((....(((........)))..))))))------. ( -29.20) >DroSim_CAF1 84890 104 + 1 AAGGCGGGCGACGGAAUGGCACUUGGUUUUUCGUUGUUCCUGGCCAUUAUUAUGGCCAUUGUUGUUGCGCCGUGCCGCUUCACACCUUUAUUUGUAUUGGUAU------U ..((((((((((((((..((............))..))))(((((((....)))))))..)))))).))))((((((....(((........)))..))))))------. ( -36.30) >DroEre_CAF1 88667 109 + 1 AAGGCGGUCGGCGGAAUGGCACUUGGUUUUUCGUUGUUCUU-GCCAUUAUAAUGGCCACUGUUGUUGCGCCGUGCCGCUUCACACCUUUAUUUGUAUUGGCAUUGGCAUU .((((((((((((.((..(((..((((....((((((....-......)))))))))).)))..)).))))).)))))))...................((....))... ( -33.80) >consensus AAGGCGGGCGACGGAAUGGCACUUGGUUUUUCGUUGUUCCU_GCCAUUAUUAUGGCCAUUGUUGUUGCGCCGUGCCGCUUCACACCUUUAUUUGUAUUGGUAU______U ..((((((((((((((..((............))..))))(((((((....)))))))..)))))).))))((((((....(((........)))..))))))....... (-28.50 = -28.88 + 0.38)

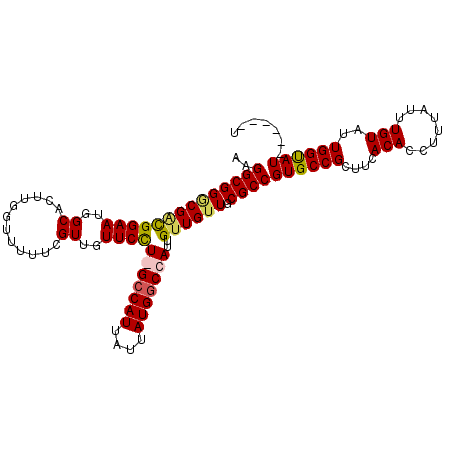

| Location | 18,854,969 – 18,855,072 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 92.04 |
| Mean single sequence MFE | -27.98 |
| Consensus MFE | -23.65 |
| Energy contribution | -24.90 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977293 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18854969 103 - 22224390 A------AUACCAAUACAAAUAAAGGUGUGAAGCGGCACGGCGCAACAACAAUGGCCAUAAUAAUGGC-AAGAACAACGAAAAACCAAGUGCCAUUCCAACGUCCGCCUU .------...............((((((.((...((...(((((..((....))(((((....)))))-...................)))))...))....)))))))) ( -23.90) >DroSec_CAF1 91534 104 - 1 A------AUACCAAUACAAAUAAAGGUGUGAAGCGGCACGGCGCAACAACAAUGGACAUAAUAAUGGCCAGGAACAACGAAAAACCAAGUGCCAUUCCGUCGCCCGCCUU .------...............((((((.(..((((...(((((........(((.(((....))).)))((............))..)))))...))))..).)))))) ( -26.60) >DroSim_CAF1 84890 104 - 1 A------AUACCAAUACAAAUAAAGGUGUGAAGCGGCACGGCGCAACAACAAUGGCCAUAAUAAUGGCCAGGAACAACGAAAAACCAAGUGCCAUUCCGUCGCCCGCCUU .------...............((((((.(..((((...(((((........(((((((....)))))))((............))..)))))...))))..).)))))) ( -33.70) >DroEre_CAF1 88667 109 - 1 AAUGCCAAUGCCAAUACAAAUAAAGGUGUGAAGCGGCACGGCGCAACAACAGUGGCCAUUAUAAUGGC-AAGAACAACGAAAAACCAAGUGCCAUUCCGCCGACCGCCUU ......................((((((....((((...(((((.......((.(((((....)))))-....))...(......)..)))))...))))....)))))) ( -27.70) >consensus A______AUACCAAUACAAAUAAAGGUGUGAAGCGGCACGGCGCAACAACAAUGGCCAUAAUAAUGGC_AAGAACAACGAAAAACCAAGUGCCAUUCCGUCGCCCGCCUU ......................((((((.(..((((...(((((........(((((((....)))))))........(......)..)))))...))))..).)))))) (-23.65 = -24.90 + 1.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:30 2006