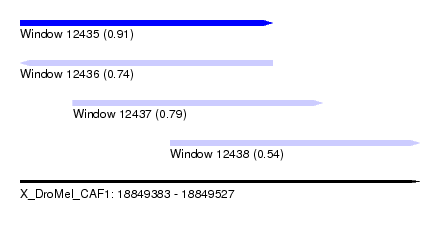
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,849,383 – 18,849,527 |
| Length | 144 |
| Max. P | 0.912635 |

| Location | 18,849,383 – 18,849,474 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 77.47 |
| Mean single sequence MFE | -28.52 |
| Consensus MFE | -17.61 |
| Energy contribution | -17.58 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
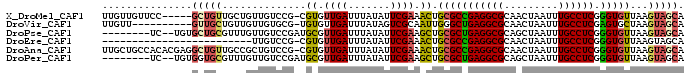
>X_DroMel_CAF1 18849383 91 + 22224390 UUGUUGUUCC-----GCUGUUGCUGUUGUCCG-CGUGUUGAUUUAUAUUCGAAACUGCGCCGAGGCGCAACUAAUUUGCCUCGGGUGUUAAGUAGCA ..........-----...((((((..(..(((-(((.((((.......))))....)))).((((((.........))))))))..)...)))))). ( -25.90) >DroVir_CAF1 3838 86 + 1 UUGUU----------GUUGCUGUUGUUGUGCG-UGUGUUGAUUUAUAGUCGCAAUUGGGCUGAGGCGCAACUAAUUUGCCUCGAGUGCUAAGUAGCA ....(----------((((((...((((((..-((((......))))..)))))).(.(((((((((.........)))))).))).)..))))))) ( -23.30) >DroPse_CAF1 118640 87 + 1 --------UC--UGUGCUGCGUUUGUUGUCCGAUGCGUUGAUUUAUAUUCGAAGCUGCGCUGAGGCGCAGCUAAUUUGCCUCGGGUGUUAAGUAGCA --------..--..((((((......(..((((.(((((((.......))))((((((((....))))))))....))).))))..)....)))))) ( -34.50) >DroEre_CAF1 83463 71 + 1 -------------------------UUGUCCG-CGUGUUGAUUUAUAUUCGAAACUGCGCCGAGGCGCAACUAAUUUGCCUCGGGUGUUAAGUAGCA -------------------------.....((-(((.((((.......)))).)).)))((((((((.........))))))))............. ( -19.50) >DroAna_CAF1 4346 96 + 1 UUGCUGCCACACGAGGCUGUUGCCGCUGUCCG-CGUGUUGAUUUAUAUUCGAAACUGCGCCGAGGCGCAACUAAUUUGCCUCGGGUGUUAAGUAGCA .(((((((((.((((((.(((((.(((.((.(-(((.((((.......))))....)))).))))))))))......)))))).)))....)))))) ( -37.90) >DroPer_CAF1 124703 87 + 1 --------UC--UGUGGUGCGUUUGUUGUCCGAUGCGUUGAUUUAUAUUCGAAGCUGCGCUGAGGCGCAGCUAAUUUGCCUCGGGUGUUAAGUAGCA --------..--..((.(((......(..((((.(((((((.......))))((((((((....))))))))....))).))))..)....))).)) ( -30.00) >consensus _________C_____GCUGCUGUUGUUGUCCG_CGUGUUGAUUUAUAUUCGAAACUGCGCCGAGGCGCAACUAAUUUGCCUCGGGUGUUAAGUAGCA ...............(((((..............((.((((.......)))).)).(((((((((((.........)))))).)))))...))))). (-17.61 = -17.58 + -0.03)

| Location | 18,849,383 – 18,849,474 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 77.47 |
| Mean single sequence MFE | -20.93 |
| Consensus MFE | -11.77 |
| Energy contribution | -11.35 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.743421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18849383 91 - 22224390 UGCUACUUAACACCCGAGGCAAAUUAGUUGCGCCUCGGCGCAGUUUCGAAUAUAAAUCAACACG-CGGACAACAGCAACAGC-----GGAACAACAA ((((.........(((((((...........)))))))(((.((...((.......))...)))-))......)))).....-----.......... ( -18.50) >DroVir_CAF1 3838 86 - 1 UGCUACUUAGCACUCGAGGCAAAUUAGUUGCGCCUCAGCCCAAUUGCGACUAUAAAUCAACACA-CGCACAACAACAGCAAC----------AACAA ((((...........(((((...........)))))........((((................-)))).......))))..----------..... ( -14.09) >DroPse_CAF1 118640 87 - 1 UGCUACUUAACACCCGAGGCAAAUUAGCUGCGCCUCAGCGCAGCUUCGAAUAUAAAUCAACGCAUCGGACAACAAACGCAGCACA--GA-------- ((((.........((((.((.....((((((((....))))))))..((.......))...)).))))...........))))..--..-------- ( -23.35) >DroEre_CAF1 83463 71 - 1 UGCUACUUAACACCCGAGGCAAAUUAGUUGCGCCUCGGCGCAGUUUCGAAUAUAAAUCAACACG-CGGACAA------------------------- .............(((((((...........)))))))(((.((...((.......))...)))-)).....------------------------- ( -14.70) >DroAna_CAF1 4346 96 - 1 UGCUACUUAACACCCGAGGCAAAUUAGUUGCGCCUCGGCGCAGUUUCGAAUAUAAAUCAACACG-CGGACAGCGGCAACAGCCUCGUGUGGCAGCAA ((((......(((.((((((.......((((((....))))))...................((-(.....)))......)))))).)))..)))). ( -32.40) >DroPer_CAF1 124703 87 - 1 UGCUACUUAACACCCGAGGCAAAUUAGCUGCGCCUCAGCGCAGCUUCGAAUAUAAAUCAACGCAUCGGACAACAAACGCACCACA--GA-------- (((..........((((.((.....((((((((....))))))))..((.......))...)).)))).........))).....--..-------- ( -22.51) >consensus UGCUACUUAACACCCGAGGCAAAUUAGUUGCGCCUCAGCGCAGUUUCGAAUAUAAAUCAACACA_CGGACAACAACAGCAGC_____G_________ ((((.(.........).))))....((((((((....)))))))).................................................... (-11.77 = -11.35 + -0.42)


| Location | 18,849,402 – 18,849,492 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 88.11 |
| Mean single sequence MFE | -27.03 |
| Consensus MFE | -23.75 |
| Energy contribution | -23.00 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18849402 90 + 22224390 G----UUGUCCGCGUGUUGAUUUAUAUUCGAAACUGCGCCGAGGCGCAACUAAUUUGCCUCGGGUGUUAAGUAGCAUGUGAAAUAAACGCAACG (----((((.(((((((((....((((((((...(((((....))))).(......)..))))))))....)))))))))........))))). ( -27.20) >DroVir_CAF1 3852 82 + 1 G----UUGUGCGUGUGUUGAUUUAUAGUCGCAAUUGGGCUGAGGCGCAACUAAUUUGCCUCGAGUGCUAAGUAGCAUGCGAAACA--------G (----((.(((((((....(((((((.((((......)).((((((.........)))))))).)).))))).))))))).))).--------. ( -24.80) >DroSec_CAF1 85971 90 + 1 G----UUGUCCGCGUGUUGAUUUAUAUUCGAAACUGCGCCGAGGCGCAACUAAUUUGCCUCGGGUGUUAAGUAGCAUGUGAAAUAAACGCAACG (----((((.(((((((((....((((((((...(((((....))))).(......)..))))))))....)))))))))........))))). ( -27.20) >DroEre_CAF1 83463 89 + 1 -----UUGUCCGCGUGUUGAUUUAUAUUCGAAACUGCGCCGAGGCGCAACUAAUUUGCCUCGGGUGUUAAGUAGCAUGCGAAAUAAACGCAACG -----.....(((((((((....((((((((...(((((....))))).(......)..))))))))....))))))))).............. ( -27.20) >DroYak_CAF1 83757 94 + 1 GUUGGUUGUCCGCGUGUUGAUUUAUAUUCGAAACUGCGCCGAGGCGCAACUAAUUUGCCUCGGGUGUUAAGUAGCAUGUGAAAUAAACGCAACG (((((((...(((((((((....((((((((...(((((....))))).(......)..))))))))....))))))))).....))).)))). ( -30.50) >DroAna_CAF1 4370 80 + 1 G----CUGUCCGCGUGUUGAUUUAUAUUCGAAACUGCGCCGAGGCGCAACUAAUUUGCCUCGGGUGUUAAGUAGCAUGUGAAAU---------- .----.....(((((((((....((((((((...(((((....))))).(......)..))))))))....)))))))))....---------- ( -25.30) >consensus G____UUGUCCGCGUGUUGAUUUAUAUUCGAAACUGCGCCGAGGCGCAACUAAUUUGCCUCGGGUGUUAAGUAGCAUGUGAAAUAAACGCAACG ..........((((((((((.......))...((((((((((((((.........)))))).)))))..))))))))))).............. (-23.75 = -23.00 + -0.75)


| Location | 18,849,437 – 18,849,527 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 72.71 |
| Mean single sequence MFE | -22.52 |
| Consensus MFE | -14.98 |
| Energy contribution | -14.48 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.20 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.541370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18849437 90 + 22224390 CGAGGCGCAACUAAUUUGCCUCGGGUGUUAAGUAGCAUGUGAAAUAAACGC----AACG-------GGA---GAAAGCCAAACAC--UCAGGAGUGUAGAGAUGGG ...(((((((.....))))((((.(((((....)))))(((.......)))----..))-------)).---....)))..((((--(....)))))......... ( -20.20) >DroVir_CAF1 3887 78 + 1 UGAGGCGCAACUAAUUUGCCUCGAGUGCUAAGUAGCAUGCGAAACA------------G-------GGAGACGACAGCCAAAAGC--AUGUGAGUGCAG------- .((((((.........))))))..(..((.....((((((......------------.-------(....)(.....)....))--)))).))..)..------- ( -21.70) >DroSec_CAF1 86006 88 + 1 CGAGGCGCAACUAAUUUGCCUCGGGUGUUAAGUAGCAUGUGAAAUAAACGC----AACG-------GGA---GAAAGCCAAACAC--UCAGGAGUGCGGUG--GGG .((((((.........))))))(((((((........((((.......)))----)...-------((.---.....)).)))))--))............--... ( -20.00) >DroEre_CAF1 83497 80 + 1 CGAGGCGCAACUAAUUUGCCUCGGGUGUUAAGUAGCAUGCGAAAUAAACGC----AACG-------GGA---GAAAGCCAAACAC--UGGAGAGUG---------- (((((((.........)))))))((((((........((((.......)))----)...-------((.---.....)).)))))--)........---------- ( -20.70) >DroYak_CAF1 83796 82 + 1 CGAGGCGCAACUAAUUUGCCUCGGGUGUUAAGUAGCAUGUGAAAUAAACGC----AACG-------GGA---GAAAGCCAAACACAUGGGAGAGUG---------- (((((((.........))))))).(((((........((((.......)))----)...-------((.---.....)).)))))...........---------- ( -17.80) >DroPer_CAF1 124753 91 + 1 UGAGGCGCAGCUAAUUUGCCUCGGGUGUUAAGUAGCAUGUGAAGUAAACGGUGGCAGCGCAGGGCAGGG---CAGAGCCC------------AGUGCCGAAAGAGA ....((((.((((.(((((.(((.(((((....))))).))).)))))...)))).))))..(((((((---(...))))------------..))))........ ( -34.70) >consensus CGAGGCGCAACUAAUUUGCCUCGGGUGUUAAGUAGCAUGUGAAAUAAACGC____AACG_______GGA___GAAAGCCAAACAC__UCGAGAGUGC_G_______ ...(((((((.....)))).(((.(((((....))))).)))..................................)))........................... (-14.98 = -14.48 + -0.50)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:28 2006