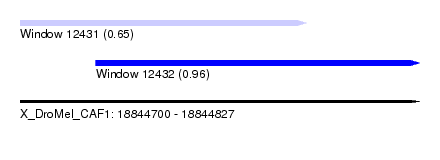
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,844,700 – 18,844,827 |
| Length | 127 |
| Max. P | 0.955221 |

| Location | 18,844,700 – 18,844,791 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.61 |
| Mean single sequence MFE | -34.35 |
| Consensus MFE | -19.67 |
| Energy contribution | -19.42 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.647810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18844700 91 + 22224390 --------------GGUGCGGGUCUUGGAGCUGUGCGG------------UGCUCAUAAUUCCAGCUUUUAU-UUGUGGGAUUUUGGCAUGGCAUGUCAGUCAUGCAUCUC--UGCAACC --------------((((((((...((.((((((((.(------------(((.((.(((((((.(......-..)))))))).)))))).)))...))))..).))..))--))).))) ( -30.00) >DroPse_CAF1 112549 118 + 1 GCGGCAGGAGGCAGGGAGCAGGGAUA-GGGGUGUGGGAUAGCUCUGGUAAAGCUCAUAAUUCCAGCUUUUAU-UUGUGGGAUUUUGGCAUGGCAUGUCAGUCAUGCACCGCAUCGCUGCC ..(((((((.((.((...((.(((((-(((((.((((((((((.......))))....))))))))))))))-)).))........(((((((......))))))).)))).)).))))) ( -42.80) >DroSec_CAF1 81308 91 + 1 --------------GGUGCGGGUCUUGGAGCUGUGCGG------------UGCUCAUAAUUCCAGCUUUUAU-UUGUGGGAUUUUGGCAUGGCAUGUCAGUCAUGCAUCUC--UGCAACC --------------((((((((...((.((((((((.(------------(((.((.(((((((.(......-..)))))))).)))))).)))...))))..).))..))--))).))) ( -30.00) >DroEre_CAF1 78777 91 + 1 -------------GGGUGCGCGUCUUGGAGCUGUGCGG------------UGCUCAUAAUUCCAGCUUUUAU-UUGUGGGAUUUUGGCAUGGCAUGUCAGUCAUGCAUCUC--UGCAUC- -------------(((((((.(.((.((....((((.(------------(((.((.(((((((.(......-..)))))))).)))))).)))).)))).).))))))).--......- ( -29.30) >DroYak_CAF1 78956 103 + 1 GAUGCGGAUG--AGGGUGCUGGUCUUGGAGCUGUGCGG------------UGCUCAUAAUUCCAGCUUUUAU-UUGUGGGAUUUUGGCAUGGCAUGUCAGUCAUGCAUCUC--UGCAUCU ((((((((..--..((((((((.((.((....((((.(------------(((.((.(((((((.(......-..)))))))).)))))).)))).))))))).)))))))--)))))). ( -39.20) >DroPer_CAF1 118500 113 + 1 GGGACAGG------GGGGCAGGGAUA-GGGGUGUGGGAUAGCUCUGGUAAAGCUCAUAAUUCCAGCUUUUAUUUUGUGGGAUUUUGGCAUGGAAUGUCAGUCAUGCACCGCACCGCUGCC .((.(((.------((....((((((-(((((.((((((((((.......))))....)))))))))))))))))((((.......((((((........)))))).)))).)).))))) ( -34.80) >consensus ______________GGUGCGGGUCUUGGAGCUGUGCGG____________UGCUCAUAAUUCCAGCUUUUAU_UUGUGGGAUUUUGGCAUGGCAUGUCAGUCAUGCAUCUC__UGCAACC ..............(((((...................................((.(((((((.(.........)))))))).))(((((((......)))))))........))).)) (-19.67 = -19.42 + -0.25)


| Location | 18,844,724 – 18,844,827 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.07 |
| Mean single sequence MFE | -26.56 |
| Consensus MFE | -18.74 |
| Energy contribution | -18.46 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18844724 103 + 22224390 ----------UGCUCAUAAUUCCAGCUUUUAU-UUGUGGGAUUUUGGCAUGGCAUGUCAGUCAUGCAUCUC--UGCAACCUC-CUUACCCGUCUCCC---UCCGCCUUCGCCUUUGCCUU ----------(((.((.(((((((.(......-..)))))))).))(((((((......))))))).....--.))).....-..............---.................... ( -19.90) >DroPse_CAF1 112588 112 + 1 GCUCUGGUAAAGCUCAUAAUUCCAGCUUUUAU-UUGUGGGAUUUUGGCAUGGCAUGUCAGUCAUGCACCGCAUCGCUGCCCC-AUGCCCCAUGCCCCAUGCCCGCAUGCGC------AUU ((......((((((.........))))))...-.((((((.....(((((((((((.((((.((((...)))).))))...)-)))))..))))).....))))))...))------... ( -38.20) >DroSec_CAF1 81332 97 + 1 ----------UGCUCAUAAUUCCAGCUUUUAU-UUGUGGGAUUUUGGCAUGGCAUGUCAGUCAUGCAUCUC--UGCAACCUC-CUUACCCAUCUCCC---UCCGCCUCCGC------CUU ----------(((.((.(((((((.(......-..)))))))).))(((((((......))))))).....--.))).....-..............---...........------... ( -19.90) >DroYak_CAF1 78992 90 + 1 ----------UGCUCAUAAUUCCAGCUUUUAU-UUGUGGGAUUUUGGCAUGGCAUGUCAGUCAUGCAUCUC--UGCAUCUUCGCCUCCGCAUCUCC-----------ACGC------CGU ----------.((.((.(((((((.(......-..)))))))).))))(((((.........(((((....--)))))....((....))......-----------..))------))) ( -20.90) >DroPer_CAF1 118533 113 + 1 GCUCUGGUAAAGCUCAUAAUUCCAGCUUUUAUUUUGUGGGAUUUUGGCAUGGAAUGUCAGUCAUGCACCGCACCGCUGCCCC-AUGCCCCAUGCCCCAUGCCCGCAUGCGC------AUU ((......((((((.........)))))).....((((((.....(((((((.(((.((((..(((...)))..))))...)-))...))))))).....))))))...))------... ( -33.90) >consensus __________UGCUCAUAAUUCCAGCUUUUAU_UUGUGGGAUUUUGGCAUGGCAUGUCAGUCAUGCAUCUC__UGCAACCUC_CUUCCCCAUCUCCC___CCCGCAUGCGC______CUU ..........(((.((.(((((((.(.........)))))))).))(((((((......)))))))........)))........................................... (-18.74 = -18.46 + -0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:23 2006