| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,088,714 – 2,088,813 |
| Length | 99 |
| Max. P | 0.515581 |

| Location | 2,088,714 – 2,088,813 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 69.17 |
| Mean single sequence MFE | -27.88 |
| Consensus MFE | -18.75 |
| Energy contribution | -18.37 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.29 |
| Mean z-score | -0.84 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.515581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2088714 99 + 22224390 UCCCUGUGCGGCGAUCUCCUGAUGGGUCGCUGGGGAAAUGUCAACCAAGGAGACUGCUUUGGUAACAACA--UAAUUUUAU-ACCUUUCC--UACAAAGAGGCU- ((((....(((((((((......))))))))))))).((((..(((((((......)))))))....)))--)........-.(((((..--.....)))))..- ( -28.10) >DroVir_CAF1 9611 96 + 1 UCGCUCUGCGGCGAUCUGCUUAUGGGUCGCUGGGGCAAUGUCAACCAGGGCGAUUGCUUCGGUAAGUGAGAUCAGAUCUCCA---GCUCCCA-GUAA-----UUA (((((((((((((((((......))))))))).(((...)))...))))))))(((((..((.....(((((...)))))..---...)).)-))))-----... ( -33.20) >DroSec_CAF1 12252 100 + 1 UCUCUGUGCGGUGAUCUCCUGAUGGGUCGCUGGGGAAAUGUCAACCAAGGAGACUGCUUUGGUAACAACA--UAACUUUAU-ACCUUACC--UGCAAAGAGGCUA (((((.((((((((((((((((....)))..))))).((((..(((((((......)))))))....)))--)........-...)))))--.))).)))))... ( -27.90) >DroWil_CAF1 8480 82 + 1 UCGCUGUGUGGUGAUCUCUUGAUGGGUCGCUGGGGCAAUGUCAAUCAGGGUGAUUGCUUUGGUAAG---------UUUUC--------CC--UAUUUU----CCA .........((((((((......))))))))((((.(((....((((((((....))))))))..)---------)).))--------))--......----... ( -20.80) >DroYak_CAF1 14845 100 + 1 UCUCUGUGCGGCGAUCUGCUGAUGGGUCGCUGGGGAAAUGUCAACCAAGGAGAUUGCUUUGGUAACAACA--UAGCUUUAU-ACUUUUAA--UGCAAAUAUGCUU ...((((((((((((((......)))))))))......(((..(((((((......))))))).))).))--)))......-........--.(((....))).. ( -26.70) >DroAna_CAF1 12581 82 + 1 UCCCUGUGCGGCGACCUGCUCAUGGGGCGCUGGGGCAAUGUCAACCAGGGCGAUUGCUUCGGUAAG---------------------UCA--UACAGACGUCCUG ....((.((((....)))).)).(((((((((((((((((((......))).)))))))))))..(---------------------((.--....)))))))). ( -30.60) >consensus UCCCUGUGCGGCGAUCUCCUGAUGGGUCGCUGGGGAAAUGUCAACCAAGGAGAUUGCUUUGGUAACAACA__UA_CUUUAU____UUUCC__UACAAA_A_GCUA ......(.(((((((((......))))))))).).........(((((((......))))))).......................................... (-18.75 = -18.37 + -0.39)

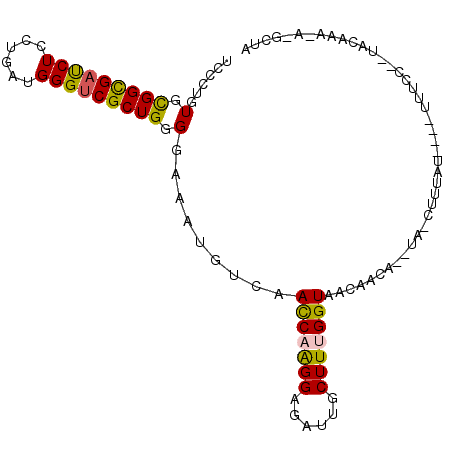
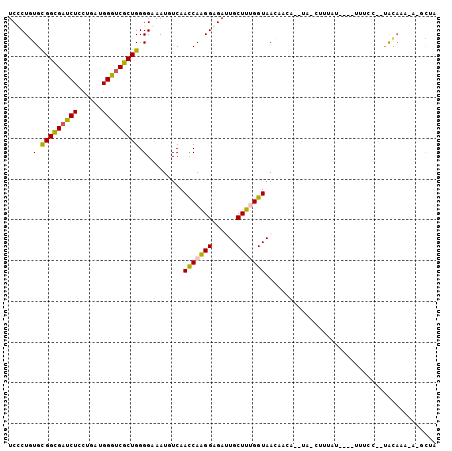
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:06 2006