
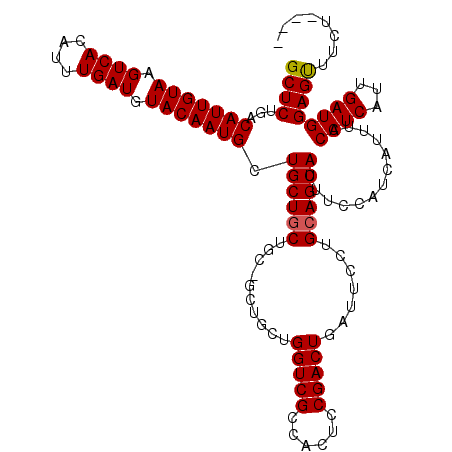
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,838,324 – 18,838,434 |
| Length | 110 |
| Max. P | 0.949669 |

| Location | 18,838,324 – 18,838,434 |
|---|---|
| Length | 110 |
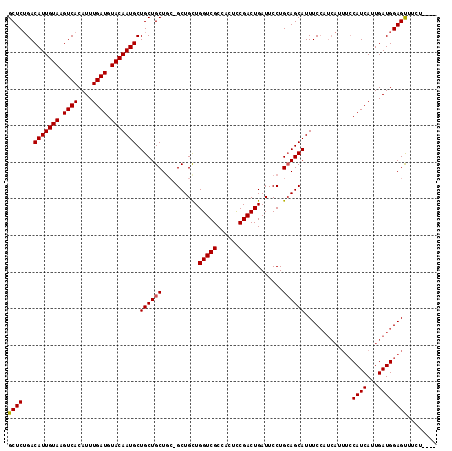
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 85.30 |
| Mean single sequence MFE | -34.77 |
| Consensus MFE | -25.72 |
| Energy contribution | -25.83 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18838324 110 + 22224390 GCUCUGACAUUGUAAGUCACAUUUGAUGUACAAUGCUGCUGCU---GCUGCUGGUCGCCACUCCGACUGAUUCCUGCAGCAUUUCCAUCAUUUCCAUCAUUGAUGGAGUUUCU---- ((...(.(((((((.((((....)))).)))))))).))((((---((.(..(((((......)))))..)....)))))).((((((((..........)))))))).....---- ( -34.40) >DroSec_CAF1 74870 113 + 1 GCUCUGACAUUGUAAGUCACAUUUGAUGUACAAUGCUGCUGCUGCCGCUGCUGGUCGCCACUCCGACUGAUUCCUGCAGCAUUUCCAUCAUUUCCAUCAUUGAUGGAGUUUCU---- ((...(.(((((((.((((....)))).)))))))).))((((((.(..(..(((((......)))))..)..).)))))).((((((((..........)))))))).....---- ( -35.60) >DroYak_CAF1 72479 106 + 1 GCUCGGACAUUGUAAGUCACAUUUGAUGUACAAUGCUGCUGCUGCUGCUGUUGGUCGCAACUUCGACUGAUUCCUGGAGCAUUUC---------CAUCAUUGAUGGAGCUA--CUAU ((.(((.(((((((.((((....)))).))))))))))..))((((.(.((..((((......))))..))....).)))).(((---------((((...)))))))...--.... ( -34.30) >consensus GCUCUGACAUUGUAAGUCACAUUUGAUGUACAAUGCUGCUGCUGC_GCUGCUGGUCGCCACUCCGACUGAUUCCUGCAGCAUUUCCAUCAUUUCCAUCAUUGAUGGAGUUUCU____ ((((...(((((((.((((....)))).))))))).((((((..........(((((......))))).......)))))).............((((...))))))))........ (-25.72 = -25.83 + 0.11)

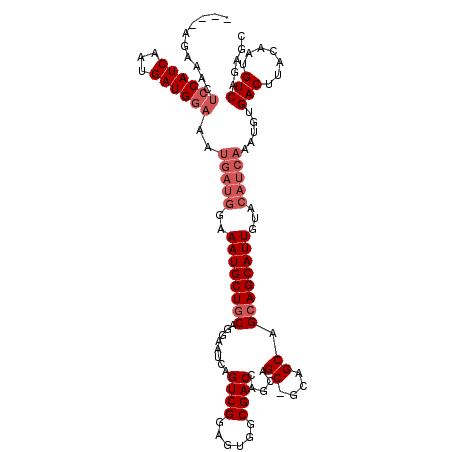
| Location | 18,838,324 – 18,838,434 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 85.30 |
| Mean single sequence MFE | -33.07 |
| Consensus MFE | -22.97 |
| Energy contribution | -25.63 |
| Covariance contribution | 2.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18838324 110 - 22224390 ----AGAAACUCCAUCAAUGAUGGAAAUGAUGGAAAUGCUGCAGGAAUCAGUCGGAGUGGCGACCAGCAGC---AGCAGCAGCAUUGUACAUCAAAUGUGACUUACAAUGUCAGAGC ----......((((((...))))))..(((((..((((((((........((((......))))..((...---.)).))))))))...)))))..(.((((.......)))).).. ( -34.90) >DroSec_CAF1 74870 113 - 1 ----AGAAACUCCAUCAAUGAUGGAAAUGAUGGAAAUGCUGCAGGAAUCAGUCGGAGUGGCGACCAGCAGCGGCAGCAGCAGCAUUGUACAUCAAAUGUGACUUACAAUGUCAGAGC ----......((((((...))))))..(((((..((((((((........((((......)))).....((....)).))))))))...)))))..(.((((.......)))).).. ( -35.60) >DroYak_CAF1 72479 106 - 1 AUAG--UAGCUCCAUCAAUGAUG---------GAAAUGCUCCAGGAAUCAGUCGAAGUUGCGACCAACAGCAGCAGCAGCAGCAUUGUACAUCAAAUGUGACUUACAAUGUCCGAGC ..((--((..((((((...))))---------))..))))...(((....((.(..(((((........)))))..).))..(((((((..(((....)))..)))))))))).... ( -28.70) >consensus ____AGAAACUCCAUCAAUGAUGGAAAUGAUGGAAAUGCUGCAGGAAUCAGUCGGAGUGGCGACCAGCAGC_GCAGCAGCAGCAUUGUACAUCAAAUGUGACUUACAAUGUCAGAGC ..........((((((...))))))..(((((..((((((((........((((......)))).....((....)).))))))))...))))).....(((.......)))..... (-22.97 = -25.63 + 2.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:17 2006