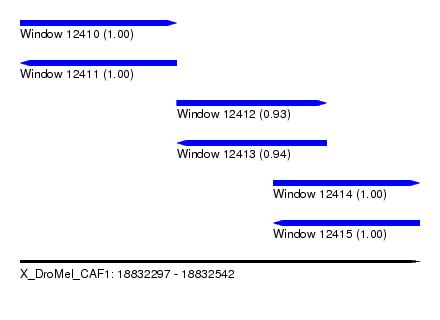
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,832,297 – 18,832,542 |
| Length | 245 |
| Max. P | 0.999989 |

| Location | 18,832,297 – 18,832,393 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -30.84 |
| Consensus MFE | -29.54 |
| Energy contribution | -29.22 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.20 |
| SVM RNA-class probability | 0.999834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18832297 96 + 22224390 ---UUUUUUUUUGGCUCAUCUCUGGCAUGCAAAUCCCGAGCCCCACACCAUCGCCCAGCAUCUUGGGCUUACUUUAUUGGCUUUUGGCUGGUGUGGGGG ---................(((.((.((....)))).)))(((((((((...((((((....))))))..........(((.....)))))))))))). ( -34.70) >DroSec_CAF1 69150 93 + 1 ------UUGUUUGGCUCAUUUCUGGCAUGCAAAUCCUGAGCCCCACACCAUCGCCCAGCAUCUUGGGCUUACUUUAUUGGCUUUUGGCUUGUGUGGGGG ------((((.((.(........).)).))))........((((((((....((((((....))))))..........(((.....))).)))))))). ( -31.20) >DroSim_CAF1 49367 96 + 1 ---UUUUUUUUUGGCUCAUCUCUGGCAUGCAAAUCCUGAGCCCCACACCAUCGCCCAGCAUCUUGGGCUUACUUUAUUGGCUUUUGGCUUGUGUGGGGG ---................(((.((.((....)))).)))((((((((....((((((....))))))..........(((.....))).)))))))). ( -29.90) >DroEre_CAF1 66883 92 + 1 -------UUUUUGGCUCUGCUCUGGCAUGCAAAUCCUGAGCCUCACACCAUCGCCCAGCAUCUUGGGCUUACUUUAUUGGCUUUUGGCUUGUGUGGGGA -------......((..(((....))).))..........((((((((....((((((....))))))..........(((.....))).)))))))). ( -27.60) >DroYak_CAF1 66001 99 + 1 UUUUUUUUUUUUGGCUCAUCUCUGGCAUGCAAAUCCUGAGCCCCACACCAUCGCCCAGGAUCUUGGGCUUACUUUAUUGGCUUUUGGCUUGUGUGGGGG ...................(((.((.((....)))).)))((((((((....((((((....))))))..........(((.....))).)))))))). ( -30.80) >consensus ___UUUUUUUUUGGCUCAUCUCUGGCAUGCAAAUCCUGAGCCCCACACCAUCGCCCAGCAUCUUGGGCUUACUUUAUUGGCUUUUGGCUUGUGUGGGGG ...................(((.((.((....)))).)))((((((((....((((((....))))))..........(((.....))).)))))))). (-29.54 = -29.22 + -0.32)

| Location | 18,832,297 – 18,832,393 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -31.36 |
| Consensus MFE | -29.56 |
| Energy contribution | -29.84 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.35 |
| Structure conservation index | 0.94 |
| SVM decision value | 5.53 |
| SVM RNA-class probability | 0.999989 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18832297 96 - 22224390 CCCCCACACCAGCCAAAAGCCAAUAAAGUAAGCCCAAGAUGCUGGGCGAUGGUGUGGGGCUCGGGAUUUGCAUGCCAGAGAUGAGCCAAAAAAAAA--- ...((((((((((.....))...........(((((......)))))..))))))))((((((...((((.....))))..)))))).........--- ( -35.70) >DroSec_CAF1 69150 93 - 1 CCCCCACACAAGCCAAAAGCCAAUAAAGUAAGCCCAAGAUGCUGGGCGAUGGUGUGGGGCUCAGGAUUUGCAUGCCAGAAAUGAGCCAAACAA------ ...((((((..((.....))...........(((((......)))))....))))))((((((...((((.....))))..))))))......------ ( -31.00) >DroSim_CAF1 49367 96 - 1 CCCCCACACAAGCCAAAAGCCAAUAAAGUAAGCCCAAGAUGCUGGGCGAUGGUGUGGGGCUCAGGAUUUGCAUGCCAGAGAUGAGCCAAAAAAAAA--- ...((((((..((.....))...........(((((......)))))....))))))((((((...((((.....))))..)))))).........--- ( -32.10) >DroEre_CAF1 66883 92 - 1 UCCCCACACAAGCCAAAAGCCAAUAAAGUAAGCCCAAGAUGCUGGGCGAUGGUGUGAGGCUCAGGAUUUGCAUGCCAGAGCAGAGCCAAAAA------- ....(((((..((.....))...........(((((......)))))....))))).(((((....((((.....))))...))))).....------- ( -25.90) >DroYak_CAF1 66001 99 - 1 CCCCCACACAAGCCAAAAGCCAAUAAAGUAAGCCCAAGAUCCUGGGCGAUGGUGUGGGGCUCAGGAUUUGCAUGCCAGAGAUGAGCCAAAAAAAAAAAA ...((((((..((.....))...........(((((......)))))....))))))((((((...((((.....))))..))))))............ ( -32.10) >consensus CCCCCACACAAGCCAAAAGCCAAUAAAGUAAGCCCAAGAUGCUGGGCGAUGGUGUGGGGCUCAGGAUUUGCAUGCCAGAGAUGAGCCAAAAAAAAA___ ...((((((..((.....))...........(((((......)))))....))))))((((((...((((.....))))..))))))............ (-29.56 = -29.84 + 0.28)


| Location | 18,832,393 – 18,832,485 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.43 |
| Mean single sequence MFE | -30.20 |
| Consensus MFE | -17.88 |
| Energy contribution | -19.35 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
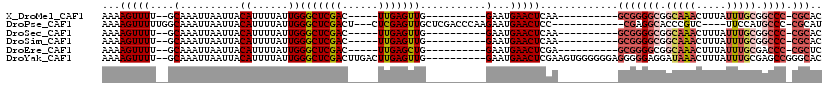
>X_DroMel_CAF1 18832393 92 + 22224390 AAAAGUUUU--GCAAAUUAAUUACAUUUUAUUGGGCUCGAC-----UUGAGUUG----------GAAUGAACUCAA----------GCGGGGCGGCAAACUUUAUUUGCGGCCC-CGCAC ..(((((..--((..(.(((.......))).)..))..)))-----))(((((.----------.....)))))..----------(((((((.(((((.....))))).))))-))).. ( -34.20) >DroPse_CAF1 93542 100 + 1 AAAAGUUUUUGGCAAAUUAAUUACAUUUUAUUGGGCUCGACU---CUCGAGUUGCUCGACCCAAGAAUGAACUCC------------CGAGGCACCCGUC----UUCCAUGCCC-CGCAU ..........((((.........(((((..(((((.(((((.---........).))))))))))))))......------------.(((((....)))----))...)))).-..... ( -21.20) >DroSec_CAF1 69243 92 + 1 AAAAGUUUU--GCAAAUUAAUUACAUUUUAUUGGGCUCGAC-----UUGAGUUG----------GAAUGAACUCAA----------GCGGGGCGGCAAACUUUAUUUGCGGCCC-CGCAC ..(((((..--((..(.(((.......))).)..))..)))-----))(((((.----------.....)))))..----------(((((((.(((((.....))))).))))-))).. ( -34.20) >DroSim_CAF1 49463 92 + 1 AAAAGUUUU--GCAAAUUAAUUACAUUUUAUUGGGCUCGAC-----UUGAGUUG----------GAAUGAACUCAA----------GCGGGGCGGCAAACUUUAUUUGCGGCCC-CGCAC ..(((((..--((..(.(((.......))).)..))..)))-----))(((((.----------.....)))))..----------(((((((.(((((.....))))).))))-))).. ( -34.20) >DroEre_CAF1 66975 92 + 1 AAAAGUUUU--GCAAAUUAAUUACAUUUUAUUGGGCUCGAC-----UUGAGCUG----------GAAUGAACUCGA----------GCGGGGCGGCAAACUUUAUUUGCGACCC-CGCUC .........--............((((((....(((((...-----..))))))----------))))).....((----------((((((..(((((.....)))))..)))-))))) ( -29.80) >DroYak_CAF1 66100 108 + 1 AAAAGUUUU--GCAAAUUAAUUACAUUUUAUUGGGCUCGACUUGACUUGAGUUG----------GAAUGAACUCGAAGUGGGGGGAGGGGGAGGAUAAACUUUAUUUGCGAGCCGGGCAC ........(--((..........((......))(((((..(((.(((((((((.----------.....))))).)))).)))(.(((..(((......)))..))).))))))..))). ( -27.60) >consensus AAAAGUUUU__GCAAAUUAAUUACAUUUUAUUGGGCUCGAC_____UUGAGUUG__________GAAUGAACUCAA__________GCGGGGCGGCAAACUUUAUUUGCGGCCC_CGCAC ...(((((....(..........((......))(((((((......)))))))...........)...))))).............(((((((.(((((.....))))).)))).))).. (-17.88 = -19.35 + 1.48)

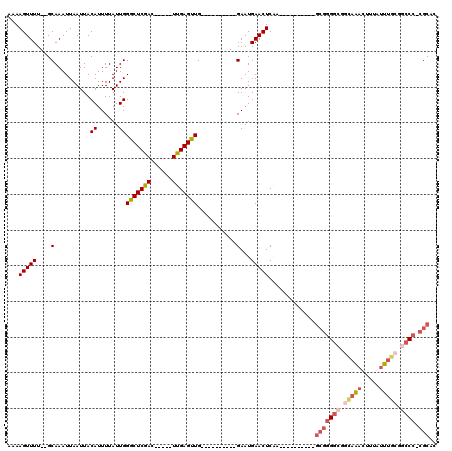
| Location | 18,832,393 – 18,832,485 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.43 |
| Mean single sequence MFE | -29.65 |
| Consensus MFE | -12.45 |
| Energy contribution | -13.34 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.66 |
| Structure conservation index | 0.42 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
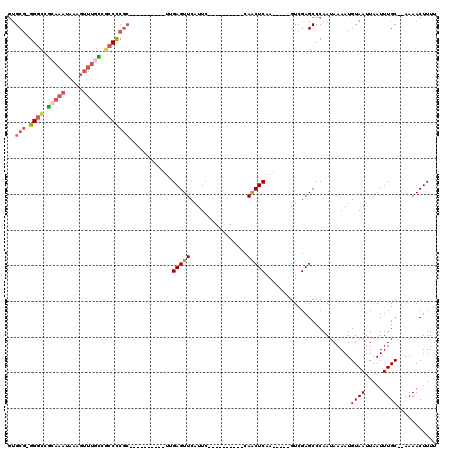
>X_DroMel_CAF1 18832393 92 - 22224390 GUGCG-GGGCCGCAAAUAAAGUUUGCCGCCCCGC----------UUGAGUUCAUUC----------CAACUCAA-----GUCGAGCCCAAUAAAAUGUAAUUAAUUUGC--AAAACUUUU (.(((-((((.((((((...)))))).)))))((----------(((((((.....----------.)))))))-----))...)).).......(((((.....))))--)........ ( -31.70) >DroPse_CAF1 93542 100 - 1 AUGCG-GGGCAUGGAA----GACGGGUGCCUCG------------GGAGUUCAUUCUUGGGUCGAGCAACUCGAG---AGUCGAGCCCAAUAAAAUGUAAUUAAUUUGCCAAAAACUUUU ...((-((((((....----.....))))))))------------((.((((((((((((((......)))))))---))).))))))........((((.....))))........... ( -30.40) >DroSec_CAF1 69243 92 - 1 GUGCG-GGGCCGCAAAUAAAGUUUGCCGCCCCGC----------UUGAGUUCAUUC----------CAACUCAA-----GUCGAGCCCAAUAAAAUGUAAUUAAUUUGC--AAAACUUUU (.(((-((((.((((((...)))))).)))))((----------(((((((.....----------.)))))))-----))...)).).......(((((.....))))--)........ ( -31.70) >DroSim_CAF1 49463 92 - 1 GUGCG-GGGCCGCAAAUAAAGUUUGCCGCCCCGC----------UUGAGUUCAUUC----------CAACUCAA-----GUCGAGCCCAAUAAAAUGUAAUUAAUUUGC--AAAACUUUU (.(((-((((.((((((...)))))).)))))((----------(((((((.....----------.)))))))-----))...)).).......(((((.....))))--)........ ( -31.70) >DroEre_CAF1 66975 92 - 1 GAGCG-GGGUCGCAAAUAAAGUUUGCCGCCCCGC----------UCGAGUUCAUUC----------CAGCUCAA-----GUCGAGCCCAAUAAAAUGUAAUUAAUUUGC--AAAACUUUU (((((-((((.((((((...)))))).)))))))----------))(((((.....----------..((((..-----...)))).........(((((.....))))--).))))).. ( -34.90) >DroYak_CAF1 66100 108 - 1 GUGCCCGGCUCGCAAAUAAAGUUUAUCCUCCCCCUCCCCCCACUUCGAGUUCAUUC----------CAACUCAAGUCAAGUCGAGCCCAAUAAAAUGUAAUUAAUUUGC--AAAACUUUU ......(((((((......((......))............((((.(((((.....----------.)))))))))...).))))))........(((((.....))))--)........ ( -17.50) >consensus GUGCG_GGGCCGCAAAUAAAGUUUGCCGCCCCGC__________UUGAGUUCAUUC__________CAACUCAA_____GUCGAGCCCAAUAAAAUGUAAUUAAUUUGC__AAAACUUUU ..(((.((((.(((((.....))))).)))))))............(((((................)))))........................((((.....))))........... (-12.45 = -13.34 + 0.89)

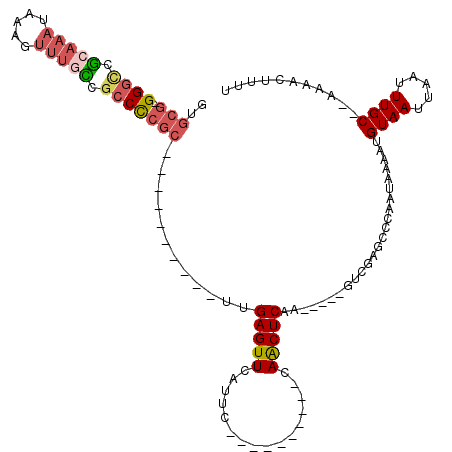
| Location | 18,832,452 – 18,832,542 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 77.24 |
| Mean single sequence MFE | -30.12 |
| Consensus MFE | -18.20 |
| Energy contribution | -19.92 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.60 |
| SVM decision value | 3.31 |
| SVM RNA-class probability | 0.998971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18832452 90 + 22224390 GCGGGGCGGCAAACUUUAUUUGCGGCCCCGCACAAUAAGCAAGGGUUUAA-AGUUCG---AGC--------------ACUUGGCCCAGUAUUAUUAUUA--AUAAUAAGC (((((((.(((((.....))))).))))))).......((..((((...(-(((...---...--------------)))).))))..((((((.....--)))))).)) ( -33.70) >DroPse_CAF1 93614 103 + 1 -CGAGGCACCCGUC----UUCCAUGCCCCGCAUAAC--UCAAGGGUUUAAAAGUUCACAGAGCGAGGGGGAGGCGAUAGAGGGGGGAGUAUUAAUAAUACUAUAAUACAU -.......(((.((----(.((.(.((((.(..(((--(....)))).....((((...))))).)))).)))....))).)))...((((((.........)))))).. ( -26.00) >DroSec_CAF1 69302 90 + 1 GCGGGGCGGCAAACUUUAUUUGCGGCCCCGCACAAUAAGCAAGGGUUUAA-AGUUCA---AGC--------------ACUUGGCCCAGUAUUAUUAAUA--AUAAUAAGC (((((((.(((((.....))))).))))))).......((..((((...(-(((...---...--------------)))).))))..((((((.....--)))))).)) ( -33.70) >DroSim_CAF1 49522 90 + 1 GCGGGGCGGCAAACUUUAUUUGCGGCCCCGCACAAUAAGCAAGGGUUUAA-AGUUCA---AGC--------------ACUUGGCCUAGUAUUAUUAAUA--AUAAUAAGC (((((((.(((((.....))))).))))))).......((..((((...(-(((...---...--------------)))).))))..((((((.....--)))))).)) ( -31.10) >DroEre_CAF1 67034 90 + 1 GCGGGGCGGCAAACUUUAUUUGCGACCCCGCUCAAUAAGCAAGGGUUUAA-AGAUCA---AGC--------------ACUUGGCCCAGUAUUAUUAAUA--AUAAUAAGC ((((((..(((((.....)))))..)))))).......((..((((...(-((....---...--------------.))).))))..((((((.....--)))))).)) ( -26.10) >consensus GCGGGGCGGCAAACUUUAUUUGCGGCCCCGCACAAUAAGCAAGGGUUUAA_AGUUCA___AGC______________ACUUGGCCCAGUAUUAUUAAUA__AUAAUAAGC (((((((.(((((.....))))).)))))))...........(((((..................................)))))........................ (-18.20 = -19.92 + 1.72)



| Location | 18,832,452 – 18,832,542 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 77.24 |
| Mean single sequence MFE | -27.36 |
| Consensus MFE | -17.62 |
| Energy contribution | -18.14 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.995832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18832452 90 - 22224390 GCUUAUUAU--UAAUAAUAAUACUGGGCCAAGU--------------GCU---CGAACU-UUAAACCCUUGCUUAUUGUGCGGGGCCGCAAAUAAAGUUUGCCGCCCCGC .........--..(((((((....(((..((((--------------...---...)))-)....)))....)))))))(((((((.((((((...)))))).))))))) ( -30.10) >DroPse_CAF1 93614 103 - 1 AUGUAUUAUAGUAUUAUUAAUACUCCCCCCUCUAUCGCCUCCCCCUCGCUCUGUGAACUUUUAAACCCUUGA--GUUAUGCGGGGCAUGGAA----GACGGGUGCCUCG- ..((((((.........))))))...(((.(((....((....((((((......(((((..........))--)))..))))))...)).)----)).))).......- ( -22.30) >DroSec_CAF1 69302 90 - 1 GCUUAUUAU--UAUUAAUAAUACUGGGCCAAGU--------------GCU---UGAACU-UUAAACCCUUGCUUAUUGUGCGGGGCCGCAAAUAAAGUUUGCCGCCCCGC ((((((((.--...))))))....(((..((((--------------...---...)))-)....)))..)).......(((((((.((((((...)))))).))))))) ( -28.90) >DroSim_CAF1 49522 90 - 1 GCUUAUUAU--UAUUAAUAAUACUAGGCCAAGU--------------GCU---UGAACU-UUAAACCCUUGCUUAUUGUGCGGGGCCGCAAAUAAAGUUUGCCGCCCCGC (((((.(((--((....))))).)))))(((..--------------((.---.(....-.......)..))...))).(((((((.((((((...)))))).))))))) ( -28.40) >DroEre_CAF1 67034 90 - 1 GCUUAUUAU--UAUUAAUAAUACUGGGCCAAGU--------------GCU---UGAUCU-UUAAACCCUUGCUUAUUGAGCGGGGUCGCAAAUAAAGUUUGCCGCCCCGC (((((.(((--((....))))).)))))(((..--------------((.---.(....-.......)..))...))).(((((((.((((((...)))))).))))))) ( -27.10) >consensus GCUUAUUAU__UAUUAAUAAUACUGGGCCAAGU______________GCU___UGAACU_UUAAACCCUUGCUUAUUGUGCGGGGCCGCAAAUAAAGUUUGCCGCCCCGC (((((.(((..........))).)))))...................................................(((((((.(((((.....))))).))))))) (-17.62 = -18.14 + 0.52)

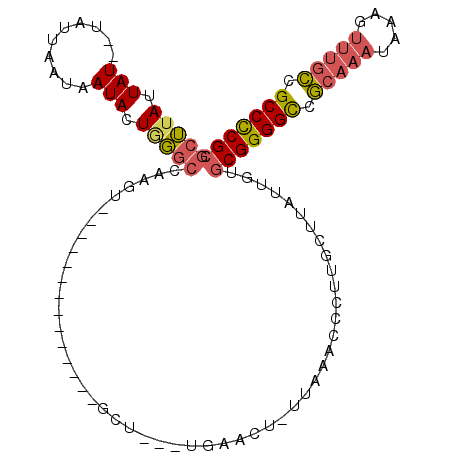
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:08 2006