
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,831,212 – 18,831,398 |
| Length | 186 |
| Max. P | 0.999242 |

| Location | 18,831,212 – 18,831,330 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 90.23 |
| Mean single sequence MFE | -36.82 |
| Consensus MFE | -31.48 |
| Energy contribution | -31.68 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.46 |
| SVM RNA-class probability | 0.999242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18831212 118 + 22224390 GAAGUGAGCUCUCGCCACUUGGCAGCAGAGUUGCAUUUAGUUGCCAUCUGCGAGGGCUUAAGUAUAUAAAUGUGCACUACCCGCAAAACAUACAUAUAGAUAGAGUAUAGUGCGAUAU ....((((((((((((....))).(((((...(((......)))..)))))))))))))).(((((....)))))......((((....((((...........))))..)))).... ( -37.90) >DroSec_CAF1 68056 116 + 1 GAAGUGAGCUCUCGCCACUUGGCAGCAGAGUUGCAUUUAGUUGCCAUCUGCGAGGGCUUAAGUAUAUAAAUGUGCACUACCCGCAA-ACAUACAUAUAGAUAGAGUAUUGUGCGAUA- ....((((((((((((....))).(((((...(((......)))..)))))))))))))).(((((....)))))......((((.-..((((...........))))..))))...- ( -38.60) >DroSim_CAF1 48284 116 + 1 GAAGUGAGCUCUCGCCACUUGGCAGCAGAGUUGCAUUUAGUUGCCAUCUGCGAGGGCUUAAGUAUAUAAAUGUGCACUACCCGCAA-ACAUACAUAUAGAUAGAGUAUAGUGCGAUA- ....((((((((((((....))).(((((...(((......)))..)))))))))))))).(((((....)))))......((((.-..((((...........))))..))))...- ( -38.60) >DroEre_CAF1 65822 109 + 1 GAAGUGAGCACUCGCCACUUGGCAGCAGAGUUGCAUUUAGUUGCCAUCUGCGAGGGCUUAAGUAUAUAAAUGUGCACUCCCCGCAA-ACACACAUAUAG------UUUAGUGCGAU-- ....(((((.((((((....))).(((((...(((......)))..)))))))).))))).(((((....)))))......((((.-....((.....)------)....))))..-- ( -32.60) >DroYak_CAF1 64862 115 + 1 GAAGUGAGCUCUCGCCACUUGGCAGCAGAGUUGCAUUUAGUUGCCAUCUGCGAGGGCUUAAGUAUAUAAAUGUGCACUUCCCGCAA-ACACACACACACACAUGUACUACUAUAGU-- ....((((((((((((....))).(((((...(((......)))..))))))))))))))(((((((...((((............-.......))))...)))))))........-- ( -36.41) >consensus GAAGUGAGCUCUCGCCACUUGGCAGCAGAGUUGCAUUUAGUUGCCAUCUGCGAGGGCUUAAGUAUAUAAAUGUGCACUACCCGCAA_ACAUACAUAUAGAUAGAGUAUAGUGCGAUA_ ....((((((((((((....))).(((((...(((......)))..)))))))))))))).(((((....)))))........................................... (-31.48 = -31.68 + 0.20)

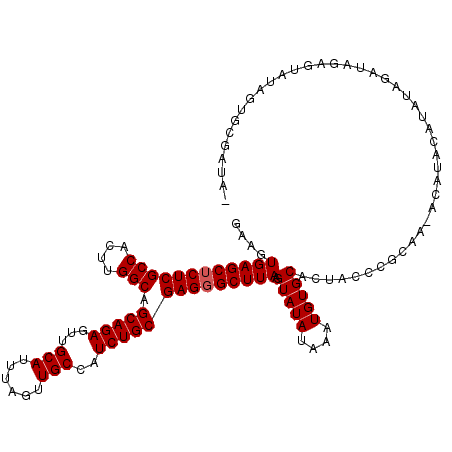
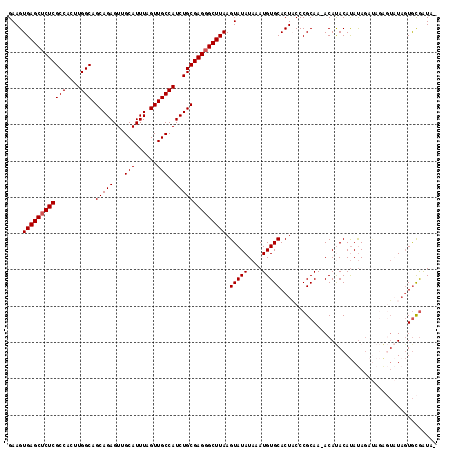
| Location | 18,831,212 – 18,831,330 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 90.23 |
| Mean single sequence MFE | -31.59 |
| Consensus MFE | -27.82 |
| Energy contribution | -27.26 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18831212 118 - 22224390 AUAUCGCACUAUACUCUAUCUAUAUGUAUGUUUUGCGGGUAGUGCACAUUUAUAUACUUAAGCCCUCGCAGAUGGCAACUAAAUGCAACUCUGCUGCCAAGUGGCGAGAGCUCACUUC .....(((((((............((((.....)))).)))))))...............(((.((((((((..(((......)))...))))).(((....)))))).)))...... ( -31.52) >DroSec_CAF1 68056 116 - 1 -UAUCGCACAAUACUCUAUCUAUAUGUAUGU-UUGCGGGUAGUGCACAUUUAUAUACUUAAGCCCUCGCAGAUGGCAACUAAAUGCAACUCUGCUGCCAAGUGGCGAGAGCUCACUUC -....((((..(((((((..(((....))).-.)).)))))))))...............(((.((((((((..(((......)))...))))).(((....)))))).)))...... ( -30.00) >DroSim_CAF1 48284 116 - 1 -UAUCGCACUAUACUCUAUCUAUAUGUAUGU-UUGCGGGUAGUGCACAUUUAUAUACUUAAGCCCUCGCAGAUGGCAACUAAAUGCAACUCUGCUGCCAAGUGGCGAGAGCUCACUUC -....(((((((............((((...-.)))).)))))))...............(((.((((((((..(((......)))...))))).(((....)))))).)))...... ( -31.52) >DroEre_CAF1 65822 109 - 1 --AUCGCACUAAA------CUAUAUGUGUGU-UUGCGGGGAGUGCACAUUUAUAUACUUAAGCCCUCGCAGAUGGCAACUAAAUGCAACUCUGCUGCCAAGUGGCGAGUGCUCACUUC --.(((((....(------(.....))....-.)))))((((((................(((.((((((((..(((......)))...))))).(((....)))))).))))))))) ( -30.19) >DroYak_CAF1 64862 115 - 1 --ACUAUAGUAGUACAUGUGUGUGUGUGUGU-UUGCGGGAAGUGCACAUUUAUAUACUUAAGCCCUCGCAGAUGGCAACUAAAUGCAACUCUGCUGCCAAGUGGCGAGAGCUCACUUC --...............(((((((.((((((-..((.....)))))))).)))))))...(((.((((((((..(((......)))...))))).(((....)))))).)))...... ( -34.70) >consensus _UAUCGCACUAUACUCUAUCUAUAUGUAUGU_UUGCGGGUAGUGCACAUUUAUAUACUUAAGCCCUCGCAGAUGGCAACUAAAUGCAACUCUGCUGCCAAGUGGCGAGAGCUCACUUC ....................((((((.((((...((.....))..)))).))))))....(((.((((((((..(((......)))...))))).(((....)))))).)))...... (-27.82 = -27.26 + -0.56)

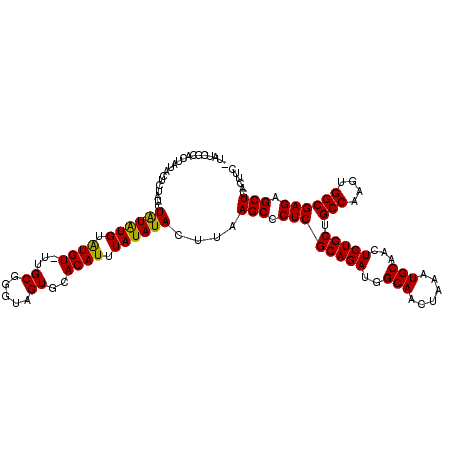

| Location | 18,831,290 – 18,831,398 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 85.58 |
| Mean single sequence MFE | -30.34 |
| Consensus MFE | -18.83 |
| Energy contribution | -18.63 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18831290 108 + 22224390 ACCCGCAAAACAUACAUAUAGAUAGAGUAUAGUGCGAUAUAUACAUAGAUGUG--UGUGGCUCCUAGGAUGCGGAUA----AAUCCAUGUGGGUAAUAUUUGACAGAAGUGAAA (((((((...(((((((((...((..(((((........))))).)).)))))--))))((((....)).))(((..----..))).)))))))....(((.((....)).))) ( -29.30) >DroSec_CAF1 68134 105 + 1 ACCCGCAA-ACAUACAUAUAGAUAGAGUAUUGUGCGAUA--UACAUAGAUGUG--UGUGGCUCCUAGGAUGCGGAUA----AAUCCAUGUGGGUAAUAUUUGACAGAAGUGAAA (((((((.-.(((((((((..((.(.(((((....))))--).)))..)))))--))))((((....)).))(((..----..))).)))))))....(((.((....)).))) ( -28.60) >DroSim_CAF1 48362 105 + 1 ACCCGCAA-ACAUACAUAUAGAUAGAGUAUAGUGCGAUA--UACAUAGAUGUG--UGUGGCUCCUAGGAUGCGGAUA----AAUCCAUGUGGGUAAUAUUUGACAGAAGUGAAA (((((((.-.(((((((((...((..(((((......))--))).)).)))))--))))((((....)).))(((..----..))).)))))))....(((.((....)).))) ( -29.50) >DroEre_CAF1 65900 95 + 1 CCCCGCAA-ACACACAUAUAG------UUUAGUGCGAU------AUAGAUGUG--UGUGGCUCCUAGGAUGCGGAUA----AAUCCAUGUGGGUAAUAUUUGACAGAAGUGAAA .((((((.-.(((((((((..------..((......)------)...)))))--))))((((....)).))(((..----..))).)))))).....(((.((....)).))) ( -25.40) >DroYak_CAF1 64940 107 + 1 UCCCGCAA-ACACACACACACACAUGUACUACUAUAGU------AUAGAUGUGUGUGUGGCUCCUAGGAUGCGGAUAAAUAAAUCCAUGUGGGUAAUAUUUGACAGAAGUGAAA .((((((.-...((((((((((..(((((((...))))------)))..))))))))))((((....)).))((((......)))).)))))).....(((.((....)).))) ( -38.90) >consensus ACCCGCAA_ACAUACAUAUAGAUAGAGUAUAGUGCGAUA__UACAUAGAUGUG__UGUGGCUCCUAGGAUGCGGAUA____AAUCCAUGUGGGUAAUAUUUGACAGAAGUGAAA .((((((...(((((((((.............................)))))..))))((((....)).))((((......)))).)))))).....(((.((....)).))) (-18.83 = -18.63 + -0.20)

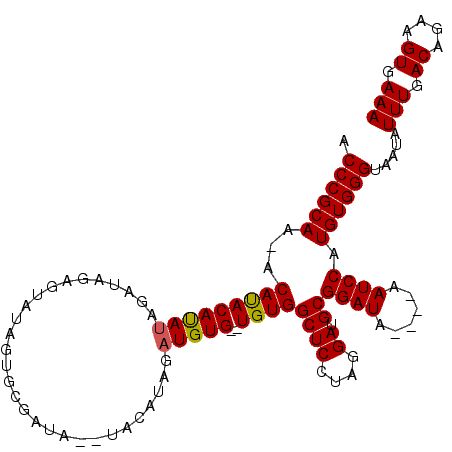
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:01 2006