
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,830,765 – 18,830,918 |
| Length | 153 |
| Max. P | 0.751670 |

| Location | 18,830,765 – 18,830,878 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.62 |
| Mean single sequence MFE | -32.75 |
| Consensus MFE | -24.20 |
| Energy contribution | -24.12 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18830765 113 + 22224390 UAUGAGCUAUUUUCC-CGAACGCCCCGCUCUGCGCUUCUCAUGGCGUUUACCUGGCUUA---UGCAACACACU---CGUCGUUUGAUUGCGGCUUAGUAGCCGUUGCAUAAUCGCUUGUC .(..(((........-.....(((.......(((((......)))))......)))(((---((((((.((.(---((.....))).)).((((....)))))))))))))..)))..). ( -33.62) >DroSec_CAF1 67615 113 + 1 UAUGAGCUAAUUUCC-CGAGCGCCCCGUUCUGCGCUUCUCAUGGCGUUUACAUCGCUUA---UGCAACACACU---CGUCGUUUGAUUGCGGCUUAGUAGCCGUUGCAUAAUCGCUUGUC ...............-((((((....((...(((((......)))))..)).....(((---((((((.((.(---((.....))).)).((((....))))))))))))).)))))).. ( -33.30) >DroSim_CAF1 47843 113 + 1 UAUGAGCUAUUUUCC-CGAGCGCCCCGCUCUGCGCUUCUCAUGGCGUUUACCUGGCUUA---UGCAACACACU---CGUCGUUUGAUUGCGGCUUAGUAGCCGUUGCAUAAUCGCUUGUC .(..(((.......(-((((((...))))).(((((......)))))......)).(((---((((((.((.(---((.....))).)).((((....)))))))))))))..)))..). ( -35.70) >DroEre_CAF1 65402 112 + 1 UAUGAGCUAUUUUC--CGAGUGCCCCUCUCCGCGCUUCUCAUGGCGUUUACCUGGCUUA---UGCAACACACU---CGUCGUUUGAAUGCGGCUUAGUUGCCAUUGCAUAAUCGCUUGUC .(..(((.......--.....(((.......(((((......)))))......)))(((---(((((..((.(---((.....))).)).(((......))).))))))))..)))..). ( -28.02) >DroYak_CAF1 64432 120 + 1 UAUGAGCUAUUUUCCCCAAGCGCCCCUCUCUGCGCUUCUCAUGGCAUUUACCUCGCUUAUGAUGCAACACACUUUGCAUCGUUUGAAUGCGGCUUAGUCGCCGUUGCAUAAUCGCUUGUC .(..(((..........((((((........))))))...(((((..(((..((((..(((((((((......)))))))))......))))..)))..))))).........)))..). ( -33.10) >consensus UAUGAGCUAUUUUCC_CGAGCGCCCCGCUCUGCGCUUCUCAUGGCGUUUACCUGGCUUA___UGCAACACACU___CGUCGUUUGAUUGCGGCUUAGUAGCCGUUGCAUAAUCGCUUGUC .(..(((..........((((((........))))))...(((((.........((.......)).....(((....(((((......)))))..))).))))).........)))..). (-24.20 = -24.12 + -0.08)



| Location | 18,830,804 – 18,830,918 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.73 |
| Mean single sequence MFE | -29.20 |
| Consensus MFE | -22.70 |
| Energy contribution | -23.10 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18830804 114 + 22224390 AUGGCGUUUACCUGGCUUA---UGCAACACACU---CGUCGUUUGAUUGCGGCUUAGUAGCCGUUGCAUAAUCGCUUGUCGCUUAAUCUAUUUAUUGAUUGACAAUAUUGAGACCCUUUA ..((.((((....((((((---((((((.((.(---((.....))).)).((((....)))))))))))))..)))(((((.(((((......))))).)))))......)))))).... ( -29.90) >DroSec_CAF1 67654 114 + 1 AUGGCGUUUACAUCGCUUA---UGCAACACACU---CGUCGUUUGAUUGCGGCUUAGUAGCCGUUGCAUAAUCGCUUGUCGCUUAAUUUAUUUAUUGAUUGACAAUAUUGAGACCCUUUA ..((.((((.((.((.(((---((((((.((.(---((.....))).)).((((....))))))))))))).)).((((((.(((((......))))).))))))...)))))))).... ( -31.50) >DroSim_CAF1 47882 114 + 1 AUGGCGUUUACCUGGCUUA---UGCAACACACU---CGUCGUUUGAUUGCGGCUUAGUAGCCGUUGCAUAAUCGCUUGUCGCUUAAUCUAUUUAUUGAUUGACAAUAUUGAGACCCUUUA ..((.((((....((((((---((((((.((.(---((.....))).)).((((....)))))))))))))..)))(((((.(((((......))))).)))))......)))))).... ( -29.90) >DroEre_CAF1 65440 114 + 1 AUGGCGUUUACCUGGCUUA---UGCAACACACU---CGUCGUUUGAAUGCGGCUUAGUUGCCAUUGCAUAAUCGCUUGUCGCUUAAUCUACUUAUUGAUUGACAAUAUUGAGACCCUUUU ..((.((((....((((((---(((((..((.(---((.....))).)).(((......))).))))))))..)))(((((.(((((......))))).)))))......)))))).... ( -25.30) >DroYak_CAF1 64472 120 + 1 AUGGCAUUUACCUCGCUUAUGAUGCAACACACUUUGCAUCGUUUGAAUGCGGCUUAGUCGCCGUUGCAUAAUCGCUUGUCGCUUAAUCUACUUAUUGAUUGACAAUAUUGAGACCCUUUA ...........(((((..(((((((((......))))))))).((...(((((......)))))..)).....))((((((.(((((......))))).))))))....)))........ ( -29.40) >consensus AUGGCGUUUACCUGGCUUA___UGCAACACACU___CGUCGUUUGAUUGCGGCUUAGUAGCCGUUGCAUAAUCGCUUGUCGCUUAAUCUAUUUAUUGAUUGACAAUAUUGAGACCCUUUA ..((.((((.....((......((((((............((......))(((......))))))))).....))((((((.(((((......))))).)))))).....)))))).... (-22.70 = -23.10 + 0.40)

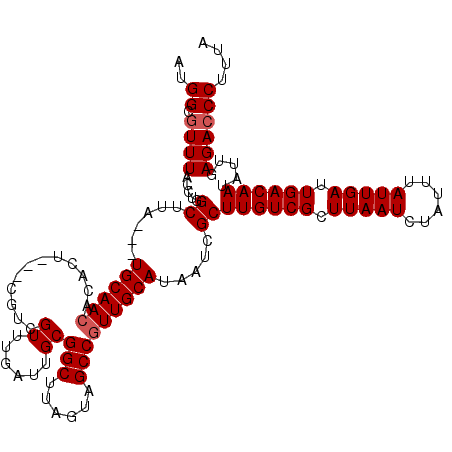

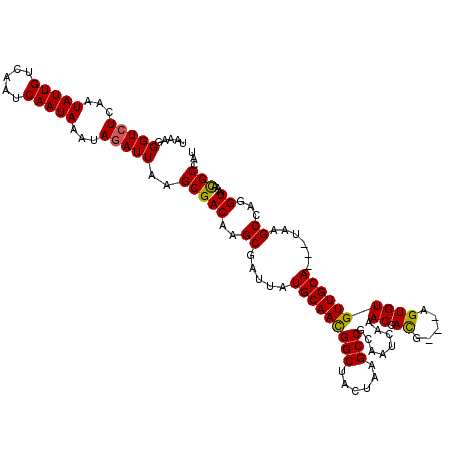
| Location | 18,830,804 – 18,830,918 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.73 |
| Mean single sequence MFE | -29.04 |
| Consensus MFE | -21.30 |
| Energy contribution | -21.02 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18830804 114 - 22224390 UAAAGGGUCUCAAUAUUGUCAAUCAAUAAAUAGAUUAAGCGACAAGCGAUUAUGCAACGGCUACUAAGCCGCAAUCAAACGACG---AGUGUGUUGCA---UAAGCCAGGUAAACGCCAU ....((((((...(((((.....)))))...))))...((.....))..(((((((((((((....))))(((.((.......)---).)))))))))---))).)).(((....))).. ( -28.60) >DroSec_CAF1 67654 114 - 1 UAAAGGGUCUCAAUAUUGUCAAUCAAUAAAUAAAUUAAGCGACAAGCGAUUAUGCAACGGCUACUAAGCCGCAAUCAAACGACG---AGUGUGUUGCA---UAAGCGAUGUAAACGCCAU .....(((....(((((((...................((.....))..(((((((((((((....))))(((.((.......)---).)))))))))---))))))))))....))).. ( -28.50) >DroSim_CAF1 47882 114 - 1 UAAAGGGUCUCAAUAUUGUCAAUCAAUAAAUAGAUUAAGCGACAAGCGAUUAUGCAACGGCUACUAAGCCGCAAUCAAACGACG---AGUGUGUUGCA---UAAGCCAGGUAAACGCCAU ....((((((...(((((.....)))))...))))...((.....))..(((((((((((((....))))(((.((.......)---).)))))))))---))).)).(((....))).. ( -28.60) >DroEre_CAF1 65440 114 - 1 AAAAGGGUCUCAAUAUUGUCAAUCAAUAAGUAGAUUAAGCGACAAGCGAUUAUGCAAUGGCAACUAAGCCGCAUUCAAACGACG---AGUGUGUUGCA---UAAGCCAGGUAAACGCCAU ....((((((...(((((.....)))))...))))...((.....))..((((((((((((......)))((((((.......)---)))))))))))---))).)).(((....))).. ( -29.00) >DroYak_CAF1 64472 120 - 1 UAAAGGGUCUCAAUAUUGUCAAUCAAUAAGUAGAUUAAGCGACAAGCGAUUAUGCAACGGCGACUAAGCCGCAUUCAAACGAUGCAAAGUGUGUUGCAUCAUAAGCGAGGUAAAUGCCAU .....((((((....(((((((((........))))....)))))((....((((((((((......)))(((((.....))))).......))))))).....)))))......))).. ( -30.50) >consensus UAAAGGGUCUCAAUAUUGUCAAUCAAUAAAUAGAUUAAGCGACAAGCGAUUAUGCAACGGCUACUAAGCCGCAAUCAAACGACG___AGUGUGUUGCA___UAAGCCAGGUAAACGCCAU .....(((((...(((((.....)))))...)))))..(((((..((.....(((((((((......)))........((.((.....))))))))))......))...))...)))... (-21.30 = -21.02 + -0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:58 2006