
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,827,087 – 18,827,362 |
| Length | 275 |
| Max. P | 0.999986 |

| Location | 18,827,087 – 18,827,202 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 65.50 |
| Mean single sequence MFE | -26.47 |
| Consensus MFE | -17.94 |
| Energy contribution | -16.83 |
| Covariance contribution | -1.10 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.995188 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18827087 115 + 22224390 CUUAGUUUUCAACUUUAACUAGCAGUGACUAUAAGGAAGCAUGAAAAAAGGUGAAAUUCCUCGCAGUGUAAGUGAGUCUCUUCGCUGGCCAUUUUAGCCCGCAAAUGGGUUUCCU ..(((((.....((......))....)))))..(((((((.........((((((....(((((.......)))))....))))))..((((((........))))))))))))) ( -25.60) >DroSec_CAF1 63926 95 + 1 CUUACUUUUC---UUACAGUUACAGUUUCU-----------------UAGGCGAAAUUUCUCGCAGUGUGAGUGAGUCCAUUCGCUGGCCAUUUUAGCCCGCAAAUGGGUUUCCU (((((((...---.(((.((...((((((.-----------------.....))))))....)).))).)))))))((((((.((.(((.......))).)).))))))...... ( -22.90) >DroEre_CAF1 61778 92 + 1 AUUUCU-----------------------UGUAAGGAAAUAUGAAAAUAGGCAAAGUUUCUCGCAGUGUGAGUGAGUCCAUUUGUUGGCCAUUUUGGCCCGCAAAUGGGUUUCCU ......-----------------------....(((((((...................(((((.......))))).((((((((.((((.....)))).))))))))))))))) ( -30.90) >consensus CUUACUUUUC___UU__A_U__CAGU__CU_UAAGGAA__AUGAAAAUAGGCGAAAUUUCUCGCAGUGUGAGUGAGUCCAUUCGCUGGCCAUUUUAGCCCGCAAAUGGGUUUCCU .................................................((((((....(((((.......)))))....)))))).........((((((....)))))).... (-17.94 = -16.83 + -1.10)



| Location | 18,827,087 – 18,827,202 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 65.50 |
| Mean single sequence MFE | -24.67 |
| Consensus MFE | -19.91 |
| Energy contribution | -20.47 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.81 |
| SVM decision value | 5.43 |
| SVM RNA-class probability | 0.999986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18827087 115 - 22224390 AGGAAACCCAUUUGCGGGCUAAAAUGGCCAGCGAAGAGACUCACUUACACUGCGAGGAAUUUCACCUUUUUUCAUGCUUCCUUAUAGUCACUGCUAGUUAAAGUUGAAAACUAAG (((.......(((((.((((.....)))).)))))((((.((.(((.......))))).)))).))).((((((.((((.....((((....))))....))))))))))..... ( -23.90) >DroSec_CAF1 63926 95 - 1 AGGAAACCCAUUUGCGGGCUAAAAUGGCCAGCGAAUGGACUCACUCACACUGCGAGAAAUUUCGCCUA-----------------AGAAACUGUAACUGUAA---GAAAAGUAAG .(....)((((((((.((((.....)))).)))))))).............(((((....)))))...-----------------..........(((....---....)))... ( -23.90) >DroEre_CAF1 61778 92 - 1 AGGAAACCCAUUUGCGGGCCAAAAUGGCCAACAAAUGGACUCACUCACACUGCGAGAAACUUUGCCUAUUUUCAUAUUUCCUUACA-----------------------AGAAAU ((((((.(((((((..((((.....))))..))))))).(((.(.......).)))....................))))))....-----------------------...... ( -26.20) >consensus AGGAAACCCAUUUGCGGGCUAAAAUGGCCAGCGAAUGGACUCACUCACACUGCGAGAAAUUUCGCCUAUUUUCAU__UUCCUUA_AG__ACUG__A_U__AA___GAAAAGUAAG (((....((((((((.((((.....)))).)))))))).(((.(.......).)))........)))................................................ (-19.91 = -20.47 + 0.56)

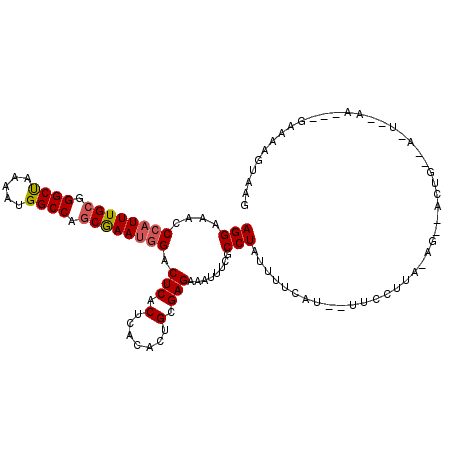

| Location | 18,827,127 – 18,827,220 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 87.10 |
| Mean single sequence MFE | -27.59 |
| Consensus MFE | -25.93 |
| Energy contribution | -24.82 |
| Covariance contribution | -1.10 |
| Combinations/Pair | 1.18 |
| Mean z-score | -0.93 |
| Structure conservation index | 0.94 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.505383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18827127 93 + 22224390 AUGAAAAAAGGUGAAAUUCCUCGCAGUGUAAGUGAGUCUCUUCGCUGGCCAUUUUAGCCCGCAAAUGGGUUUCCUCGUUUCCUUGGCCGCGUG .........((((((....(((((.......)))))....))))))(((((....((((((....))))))............)))))..... ( -24.19) >DroSec_CAF1 63953 86 + 1 -------UAGGCGAAAUUUCUCGCAGUGUGAGUGAGUCCAUUCGCUGGCCAUUUUAGCCCGCAAAUGGGUUUCCUCGUUUCCUUGGCCGCGUG -------..((((((....(((((.......)))))....))))))(((((....((((((....))))))............)))))..... ( -26.79) >DroEre_CAF1 61795 93 + 1 AUGAAAAUAGGCAAAGUUUCUCGCAGUGUGAGUGAGUCCAUUUGUUGGCCAUUUUGGCCCGCAAAUGGGUUUCCUCGUUUCCUUGGCCGCGUG .........(((.(((...(((((...))))).((((((((((((.((((.....)))).)))))))))....))).....))).)))..... ( -31.80) >consensus AUGAAAAUAGGCGAAAUUUCUCGCAGUGUGAGUGAGUCCAUUCGCUGGCCAUUUUAGCCCGCAAAUGGGUUUCCUCGUUUCCUUGGCCGCGUG .........((((((....(((((.......)))))....))))))(((((....((((((....))))))............)))))..... (-25.93 = -24.82 + -1.10)


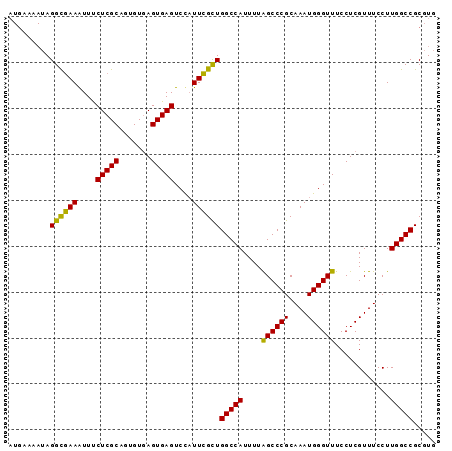
| Location | 18,827,127 – 18,827,220 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 87.10 |
| Mean single sequence MFE | -28.80 |
| Consensus MFE | -27.29 |
| Energy contribution | -27.63 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18827127 93 - 22224390 CACGCGGCCAAGGAAACGAGGAAACCCAUUUGCGGGCUAAAAUGGCCAGCGAAGAGACUCACUUACACUGCGAGGAAUUUCACCUUUUUUCAU ...((((....(....)(((........(((((.((((.....)))).)))))....))).......))))((((((........)))))).. ( -23.70) >DroSec_CAF1 63953 86 - 1 CACGCGGCCAAGGAAACGAGGAAACCCAUUUGCGGGCUAAAAUGGCCAGCGAAUGGACUCACUCACACUGCGAGAAAUUUCGCCUA------- ..(((((....(....)(((((...((((((((.((((.....)))).))))))))..)).)))...)))))..............------- ( -31.10) >DroEre_CAF1 61795 93 - 1 CACGCGGCCAAGGAAACGAGGAAACCCAUUUGCGGGCCAAAAUGGCCAACAAAUGGACUCACUCACACUGCGAGAAACUUUGCCUAUUUUCAU ..(((((....(....)(((((...(((((((..((((.....))))..)))))))..)).)))...)))))..................... ( -31.60) >consensus CACGCGGCCAAGGAAACGAGGAAACCCAUUUGCGGGCUAAAAUGGCCAGCGAAUGGACUCACUCACACUGCGAGAAAUUUCGCCUAUUUUCAU ..(((((....(....)(((((...((((((((.((((.....)))).))))))))..)).)))...)))))..................... (-27.29 = -27.63 + 0.34)



| Location | 18,827,220 – 18,827,322 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 97.39 |
| Mean single sequence MFE | -37.60 |
| Consensus MFE | -36.81 |
| Energy contribution | -37.03 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18827220 102 - 22224390 CUUGCGUGCCUCUUGCGGGAAUGGCUGUUGGCGGUUUCGCUUUCAGCGUUUUAUUUCAAUGCGAUUGAAAGCAAAUUGAAAGCCGGCACCUGCCAGAGAGCA ......(((((((.(((((..(((((.....((((((.(((((((((((...........))).))))))))))))))..)))))...))))).)))).))) ( -40.10) >DroSec_CAF1 64039 102 - 1 CUUGCGUGCCUCUUGCGGGAAUGGCUGUUGGCGGUUUCGCUUUCAGCGUUUUAUUUCAAUGCGAUUGAAAGCAAAUUGAAAGCCGGCACCUGCCAGAGAGCA ......(((((((.(((((..(((((.....((((((.(((((((((((...........))).))))))))))))))..)))))...))))).)))).))) ( -40.10) >DroEre_CAF1 61888 102 - 1 CUUGCGUGCCACUUGCGGCAACGGCUGUUGGCGGUUUCGCUUUUAGCGUUUUAUUUCAAUGCGAUUGAAAGCAAAUUGAAAGCCGGCACCUGCCAGAGAGCA ....((((((......))).)))(((.((((((((.(((((((((..((((..(((((((...)))))))..)))))))))).))).))).)))))..))). ( -32.60) >consensus CUUGCGUGCCUCUUGCGGGAAUGGCUGUUGGCGGUUUCGCUUUCAGCGUUUUAUUUCAAUGCGAUUGAAAGCAAAUUGAAAGCCGGCACCUGCCAGAGAGCA ......(((((((.(((((..(((((.....((((((.(((((((((((...........))).))))))))))))))..)))))...))))).)))).))) (-36.81 = -37.03 + 0.23)



| Location | 18,827,242 – 18,827,362 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -34.57 |
| Consensus MFE | -29.63 |
| Energy contribution | -29.63 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.86 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513170 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18827242 120 - 22224390 CCCCCCCCCCCCAUCAAAUCCAUCAGCCGUCCCUGUCGCACUUGCGUGCCUCUUGCGGGAAUGGCUGUUGGCGGUUUCGCUUUCAGCGUUUUAUUUCAAUGCGAUUGAAAGCAAAUUGAA ...................(((.(((((((.(((((.((((....)))).....))))).))))))).)))((((((.(((((((((((...........))).)))))))))))))).. ( -40.50) >DroSec_CAF1 64061 120 - 1 CAAAAGCCCCCCAUCCCAUUCAUCAGCUCUCCCUGUCGCACUUGCGUGCCUCUUGCGGGAAUGGCUGUUGGCGGUUUCGCUUUCAGCGUUUUAUUUCAAUGCGAUUGAAAGCAAAUUGAA ...................(((.(((((.((((.(..((((....))))..)....))))..))))).)))((((((.(((((((((((...........))).)))))))))))))).. ( -33.70) >DroEre_CAF1 61910 118 - 1 CAAAAG--CCCCAUCCCAUCCAUCACCUCUCCCUGUCGCUCUUGCGUGCCACUUGCGGCAACGGCUGUUGGCGGUUUCGCUUUUAGCGUUUUAUUUCAAUGCGAUUGAAAGCAAAUUGAA ((((((--(((((...((.((...............(((....)))((((......))))..)).)).))).))))).(((((((((((...........))).))))))))...))).. ( -29.50) >consensus CAAAAGCCCCCCAUCCCAUCCAUCAGCUCUCCCUGUCGCACUUGCGUGCCUCUUGCGGGAAUGGCUGUUGGCGGUUUCGCUUUCAGCGUUUUAUUUCAAUGCGAUUGAAAGCAAAUUGAA ...................(((.(((((.((((...(((....))).((.....))))))..))))).)))((((((.(((((((((((...........))).)))))))))))))).. (-29.63 = -29.63 + 0.01)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:54 2006