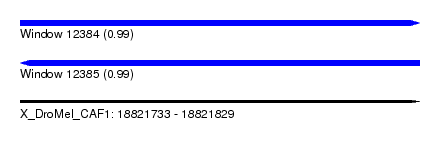
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,821,733 – 18,821,829 |
| Length | 96 |
| Max. P | 0.994890 |

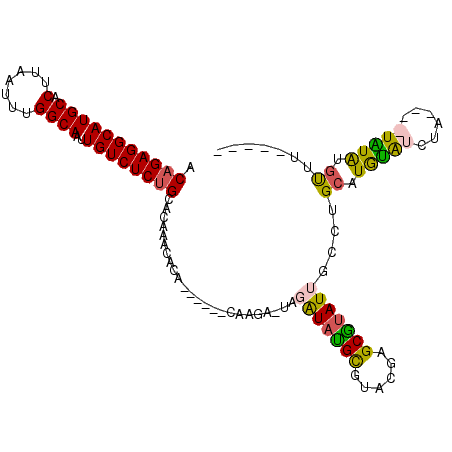
| Location | 18,821,733 – 18,821,829 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 77.56 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -16.48 |
| Energy contribution | -17.72 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.56 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18821733 96 + 22224390 -----AAACAUAUA----UAGAUACAUGCAGGCAAUACGCUCGUACGCAUAUCUA-UCUUG------UGUGUUUGUGCCGAGACAAUUGCCAAAUUAAGUGCAUGCCUCUGU -----........(----((((..((((((((((((...(((((((((((((...-....)------)))))..))).))))...))))))........))))))..))))) ( -27.80) >DroSec_CAF1 58754 101 + 1 CACCCAAACGCAUA----UAGAUACAUGCAGGCAAUAUGCUCGUACGCAUAUCUA-UCUUG------UGUGUUUGUGCAGAGACAAUUGCCAAAUUAAGUGCAUGCCUCUGU ....((((((((((----.(((((..((....))((((((......)))))).))-)))))------)))))))).((((((.((..((((.......).))))).)))))) ( -28.70) >DroEre_CAF1 56755 107 + 1 ----UAUGUAUACAUGUAUAAACAUAAGCAUGCAAUACGCA-AUACACGUACACACGCUUGUGCGUUUGUGUUUGUGCAGAGACAAUUGCCAAAUUAAGUGCAUGCCUCUGU ----...........((((((((((((((.(((.....)))-......(((((......)))))))))))))))))))((((.((..((((.......).))))).)))).. ( -29.70) >DroYak_CAF1 57720 111 + 1 CACAGAAACAUACAUAAAUACAUGCAGGCAGGCAAUACGCUCGUACGCAAACAUA-UCUUGUGCGUUUGUGUUUGUGCAGAGACAAUUGCCAAAUUAAGUGCAUGCCUCUGU .(((((..(((.(((.......((....))((((((...(((...((((((((((-...........))))))))))..)))...)))))).......))).)))..))))) ( -31.30) >consensus _____AAACAUACA____UAGAUACAUGCAGGCAAUACGCUCGUACGCAUACAUA_UCUUG______UGUGUUUGUGCAGAGACAAUUGCCAAAUUAAGUGCAUGCCUCUGU ..................((((..((((((((((((...(((...((((.(((((............))))).))))..)))...))))))........))))))..)))). (-16.48 = -17.72 + 1.25)

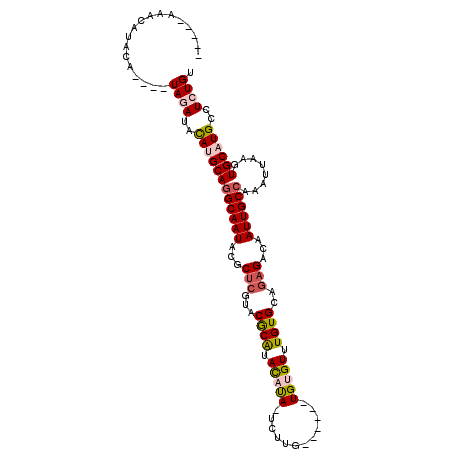

| Location | 18,821,733 – 18,821,829 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 77.56 |
| Mean single sequence MFE | -31.65 |
| Consensus MFE | -17.40 |
| Energy contribution | -17.15 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.994890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18821733 96 - 22224390 ACAGAGGCAUGCACUUAAUUUGGCAAUUGUCUCGGCACAAACACA------CAAGA-UAGAUAUGCGUACGAGCGUAUUGCCUGCAUGUAUCUA----UAUAUGUUU----- ..(((.(((((((........(((...(((((.(...........------).)))-))(((((((......))))))))))))))))).))).----.........----- ( -24.50) >DroSec_CAF1 58754 101 - 1 ACAGAGGCAUGCACUUAAUUUGGCAAUUGUCUCUGCACAAACACA------CAAGA-UAGAUAUGCGUACGAGCAUAUUGCCUGCAUGUAUCUA----UAUGCGUUUGGGUG .(((((((((((.(.......))))..))))))))..(((((...------..((.-..(((((((......)))))))..))((((((....)----)))))))))).... ( -31.70) >DroEre_CAF1 56755 107 - 1 ACAGAGGCAUGCACUUAAUUUGGCAAUUGUCUCUGCACAAACACAAACGCACAAGCGUGUGUACGUGUAU-UGCGUAUUGCAUGCUUAUGUUUAUACAUGUAUACAUA---- .(((((((((((.(.......))))..))))))))...........((((....))))((((((((((((-.(((((..(....).)))))..))))))))))))...---- ( -35.40) >DroYak_CAF1 57720 111 - 1 ACAGAGGCAUGCACUUAAUUUGGCAAUUGUCUCUGCACAAACACAAACGCACAAGA-UAUGUUUGCGUACGAGCGUAUUGCCUGCCUGCAUGUAUUUAUGUAUGUUUCUGUG (((((((((((((........((((((((.((((((.((((((....(......).-..)))))).))).))))).))))))((....))........))))))))))))). ( -35.00) >consensus ACAGAGGCAUGCACUUAAUUUGGCAAUUGUCUCUGCACAAACACA______CAAGA_UAGAUAUGCGUACGAGCGUAUUGCCUGCAUGUAUCUA____UAUAUGUUU_____ .(((((((((((.(.......))))..))))))))........................(((((((......)))))))....((.(((((......))))).))....... (-17.40 = -17.15 + -0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:42 2006