
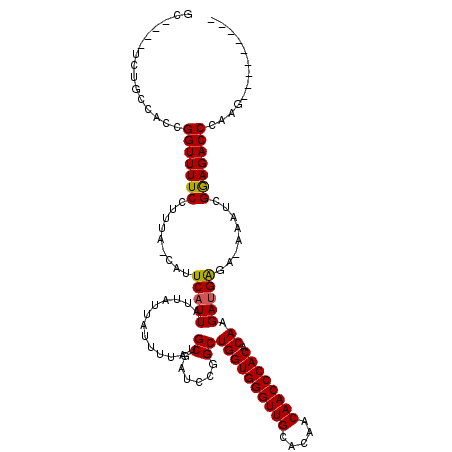
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,819,372 – 18,819,487 |
| Length | 115 |
| Max. P | 0.969533 |

| Location | 18,819,372 – 18,819,487 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 88.56 |
| Mean single sequence MFE | -31.09 |
| Consensus MFE | -24.43 |
| Energy contribution | -24.23 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18819372 115 + 22224390 GC----UCUGCCACCGGUUUUCCUUUAUCAUUCAUUAUUAUUAUUUUAUGCGAUCCGGCUGGUGGGUUGCACAACAACCCACACAAGAUGACAAAAAUCGAAGACCCAAAAGCAGCAAG ..----.((((....(((((((.........(((((.............((......))((((((((((.....)))))))).)).)))))........))))))).....)))).... ( -28.83) >DroSec_CAF1 56440 105 + 1 GC----UCUGCCACCGGUUUUCCUUUAGCAUUCAUUAUUAUUAUUUUAUGCGAUCCGGCUGGUGGGUUGCACAACAACCCACACAAGAUGAGA-AAAUCGGAGACCCAAG--------- ..----.......(((((((((.....((((....((....))....))))..((...((.((((((((.....))))))))...))..))))-))))))).........--------- ( -29.00) >DroSim_CAF1 41687 105 + 1 GC----UCUGCCACCGGUUUUCCUUUAGCAUUCAUUAUUAUUAUUUUAUGCGAUCCGGCUGGUGGGUUGCACAACAACCCACACAAGAUGAGA-AAAUCGGAGACCCAAG--------- ..----.......(((((((((.....((((....((....))....))))..((...((.((((((((.....))))))))...))..))))-))))))).........--------- ( -29.00) >DroEre_CAF1 54566 100 + 1 GC----UCUGCCACCGGUUUUCCUUUA----UCAUUAUUAUUAUUUUAUGCGAUCCGGCUGGUGGGUUGCACAACAACCCACACAAGAUGGGA-AAAUCGGAGACCG-AG--------- .(----((.(.(.(((((((((((..(----((................((......))((((((((((.....)))))))).)).)))))))-))))))).).).)-))--------- ( -34.00) >DroYak_CAF1 55377 104 + 1 GCUCGCUCAGCCACCGGUUUUCCUUUA----UCAUUAUUAUUAUUUUAUGCCAUCCGGCUGGUGGGUUGCACAACAACCCACACAAGACGGGA-AAAUCGGAGACCG-AA--------- ..(((.((.....(((((((((((...----((................(((....)))((((((((((.....)))))))).)).)).))))-))))))).)).))-).--------- ( -34.60) >consensus GC____UCUGCCACCGGUUUUCCUUUA_CAUUCAUUAUUAUUAUUUUAUGCGAUCCGGCUGGUGGGUUGCACAACAACCCACACAAGAUGAGA_AAAUCGGAGACCCAAG_________ ...............(((((((.........(((((.............((......))((((((((((.....)))))))).)).)))))........)))))))............. (-24.43 = -24.23 + -0.20)


| Location | 18,819,372 – 18,819,487 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 88.56 |
| Mean single sequence MFE | -34.32 |
| Consensus MFE | -24.78 |
| Energy contribution | -24.58 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18819372 115 - 22224390 CUUGCUGCUUUUGGGUCUUCGAUUUUUGUCAUCUUGUGUGGGUUGUUGUGCAACCCACCAGCCGGAUCGCAUAAAAUAAUAAUAAUGAAUGAUAAAGGAAAACCGGUGGCAGA----GC ....(((((..(.(((.(((...(((((((((..((.((((((((.....))))))))))((......))..................)))))))))))).))).).))))).----.. ( -34.00) >DroSec_CAF1 56440 105 - 1 ---------CUUGGGUCUCCGAUUU-UCUCAUCUUGUGUGGGUUGUUGUGCAACCCACCAGCCGGAUCGCAUAAAAUAAUAAUAAUGAAUGCUAAAGGAAAACCGGUGGCAGA----GC ---------(((..(((.(((.(((-(((.((((.((((((((((.....))))))))..)).)))).((((................))))....)))))).))).))).))----). ( -31.89) >DroSim_CAF1 41687 105 - 1 ---------CUUGGGUCUCCGAUUU-UCUCAUCUUGUGUGGGUUGUUGUGCAACCCACCAGCCGGAUCGCAUAAAAUAAUAAUAAUGAAUGCUAAAGGAAAACCGGUGGCAGA----GC ---------(((..(((.(((.(((-(((.((((.((((((((((.....))))))))..)).)))).((((................))))....)))))).))).))).))----). ( -31.89) >DroEre_CAF1 54566 100 - 1 ---------CU-CGGUCUCCGAUUU-UCCCAUCUUGUGUGGGUUGUUGUGCAACCCACCAGCCGGAUCGCAUAAAAUAAUAAUAAUGA----UAAAGGAAAACCGGUGGCAGA----GC ---------((-(.(((.(((.(((-(((.(((.((.((((((((.....))))))))))((......))................))----)...)))))).))).))).))----). ( -34.10) >DroYak_CAF1 55377 104 - 1 ---------UU-CGGUCUCCGAUUU-UCCCGUCUUGUGUGGGUUGUUGUGCAACCCACCAGCCGGAUGGCAUAAAAUAAUAAUAAUGA----UAAAGGAAAACCGGUGGCUGAGCGAGC ---------((-(((((.(((.(((-(((.(((.((.((((((((.....))))))))))(((....)))................))----)...)))))).))).)))))))..... ( -39.70) >consensus _________CUUGGGUCUCCGAUUU_UCUCAUCUUGUGUGGGUUGUUGUGCAACCCACCAGCCGGAUCGCAUAAAAUAAUAAUAAUGAAUG_UAAAGGAAAACCGGUGGCAGA____GC ..............(((.(((.(((.(((.....((.((((((((.....))))))))))((......))..........................)))))).))).)))......... (-24.78 = -24.58 + -0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:38 2006