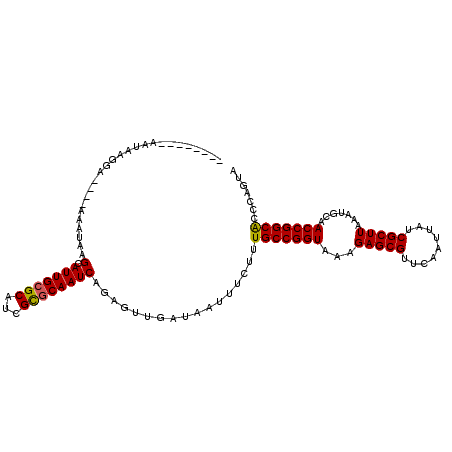
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,810,799 – 18,810,904 |
| Length | 105 |
| Max. P | 0.944226 |

| Location | 18,810,799 – 18,810,904 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 89.79 |
| Mean single sequence MFE | -26.10 |
| Consensus MFE | -23.44 |
| Energy contribution | -23.44 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.923607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18810799 105 + 22224390 UACUGGGUGCCGGUUGCAUUUAAGCGAUAAUUGAACGCUCUUUACCGGCAAAGAAAUUAUCAACUCUGAUUGCGCGAUGCGCAAUGCUUAUUUUCUUUCCUUAUU-------- ....((((((((((.((.(((((.......))))).)).....)))))))((((((.(((((....)))((((((...))))))....)).))))))))).....-------- ( -26.80) >DroSec_CAF1 48242 101 + 1 UACUGGGCGCCGGUUGCAUUUAAGCGAUAAUUGAACGCUCUUUACCGGCAAAGAAAUUAUCAACUCUGAUUGCACGAUGCGCAAUGCUUAUUU----UCCUUAUU-------- ...(((((((((((.((.(((((.......))))).)).....))))))...................(((((.......))))))))))...----........-------- ( -22.90) >DroSim_CAF1 32204 101 + 1 UACUGGGCGCCGGUUGCAUUUAAGCGAUAAUUGAACGCUCUUUACCGGCAAAGAAAUUAUCAACUCUGAUUGCGCGAUGCGCAAUGCUUAUUU----UCCUUAUU-------- ...(((((((((((.((.(((((.......))))).)).....))))))...................(((((((...))))))))))))...----........-------- ( -27.40) >DroEre_CAF1 46457 109 + 1 UACUGGUUGCCGGUUGCAUUUAAGCGAUAAUUGAACGCUCUUUACCGGCAAGGAAAUUAUCAACUCUGAUUGCGCGAUGCGCAAUGCUUAUUU----GUCUUAUUUUCCUUUU ......((((((((.((.(((((.......))))).)).....))))))))(((((.((.(((....((((((((...))))))).)....))----)...)).))))).... ( -30.30) >DroYak_CAF1 46974 93 + 1 UACUGGGUGCCGGUUGCAUUUAAGCGAUAAUUGAACGCUCUUUACCGGCAAAGAAAUUAUCAACUCUGAUUGUGCGAUGCGCAAUGCUUAUUU-------------------- ..((...(((((((.((.(((((.......))))).)).....))))))).))...............(((((((...)))))))........-------------------- ( -23.10) >consensus UACUGGGUGCCGGUUGCAUUUAAGCGAUAAUUGAACGCUCUUUACCGGCAAAGAAAUUAUCAACUCUGAUUGCGCGAUGCGCAAUGCUUAUUU____UCCUUAUU________ ...(((((((((((.((.(((((.......))))).)).....))))))...................(((((((...))))))))))))....................... (-23.44 = -23.44 + -0.00)

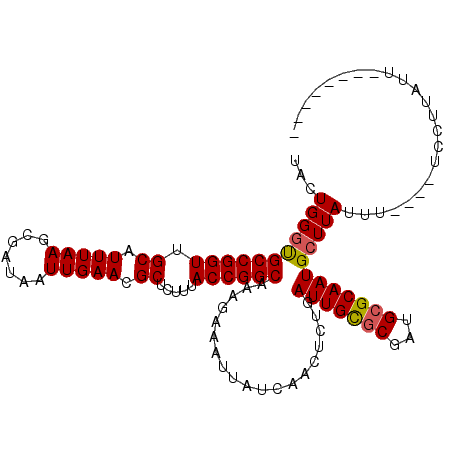
| Location | 18,810,799 – 18,810,904 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 89.79 |
| Mean single sequence MFE | -25.20 |
| Consensus MFE | -21.84 |
| Energy contribution | -21.64 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.944226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18810799 105 - 22224390 --------AAUAAGGAAAGAAAAUAAGCAUUGCGCAUCGCGCAAUCAGAGUUGAUAAUUUCUUUGCCGGUAAAGAGCGUUCAAUUAUCGCUUAAAUGCAACCGGCACCCAGUA --------.....(((((((((...((((((((((...)))))))....))).....)))))))((((((...(((((.........))))).......))))))..)).... ( -28.40) >DroSec_CAF1 48242 101 - 1 --------AAUAAGGA----AAAUAAGCAUUGCGCAUCGUGCAAUCAGAGUUGAUAAUUUCUUUGCCGGUAAAGAGCGUUCAAUUAUCGCUUAAAUGCAACCGGCGCCCAGUA --------...(((((----((...((((((((((...)))))))....))).....)))))))((((((...(((((.........))))).......))))))........ ( -24.00) >DroSim_CAF1 32204 101 - 1 --------AAUAAGGA----AAAUAAGCAUUGCGCAUCGCGCAAUCAGAGUUGAUAAUUUCUUUGCCGGUAAAGAGCGUUCAAUUAUCGCUUAAAUGCAACCGGCGCCCAGUA --------...(((((----((...((((((((((...)))))))....))).....)))))))((((((...(((((.........))))).......))))))........ ( -25.90) >DroEre_CAF1 46457 109 - 1 AAAAGGAAAAUAAGAC----AAAUAAGCAUUGCGCAUCGCGCAAUCAGAGUUGAUAAUUUCCUUGCCGGUAAAGAGCGUUCAAUUAUCGCUUAAAUGCAACCGGCAACCAGUA ..(((((((.((...(----((....(.(((((((...))))))))....))).)).)))))))((((((...(((((.........))))).......))))))........ ( -29.20) >DroYak_CAF1 46974 93 - 1 --------------------AAAUAAGCAUUGCGCAUCGCACAAUCAGAGUUGAUAAUUUCUUUGCCGGUAAAGAGCGUUCAAUUAUCGCUUAAAUGCAACCGGCACCCAGUA --------------------......(.((((.((...)).)))))((((........)))).(((((((...(((((.........))))).......)))))))....... ( -18.50) >consensus ________AAUAAGGA____AAAUAAGCAUUGCGCAUCGCGCAAUCAGAGUUGAUAAUUUCUUUGCCGGUAAAGAGCGUUCAAUUAUCGCUUAAAUGCAACCGGCACCCAGUA ..........................(.(((((((...)))))))).................(((((((...(((((.........))))).......)))))))....... (-21.84 = -21.64 + -0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:24 2006