
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,803,388 – 18,803,492 |
| Length | 104 |
| Max. P | 0.999522 |

| Location | 18,803,388 – 18,803,492 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 76.43 |
| Mean single sequence MFE | -14.97 |
| Consensus MFE | -10.86 |
| Energy contribution | -10.68 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888706 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
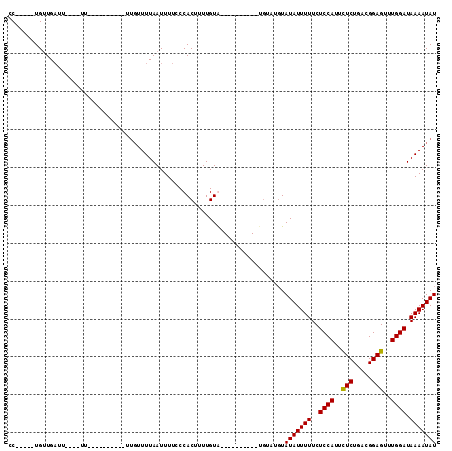
>X_DroMel_CAF1 18803388 104 + 22224390 CC-----UGUUGAUU----UUUUUUUUGUUUUUGUUUUAAUUUUCCCACUUUUGUAUAUACGAGUAUGUGUGUAUAUUUUUCUCCAUUCUCUGACGGAGUUUGGAUAAAAUAU ..-----........----.............(((((((.............(((((((((......)))))))))......((((..(((.....)))..))))))))))). ( -18.10) >DroSec_CAF1 40956 84 + 1 CC-----UGUUGAUU----UU----------UUGUUUUAAUUUUCCCACUUUUGUG----------UGUAUGUAUAUUUUUCUCCAUUCUCUGACGGAGUUUGGAUAAAAUAU ..-----........----..----------.(((((((.......(((......)----------))..............((((..(((.....)))..))))))))))). ( -13.20) >DroSim_CAF1 24245 84 + 1 CC-----UGUUGAUU----UU----------UUGUUUUAAUUUUCCCACUUUUGUG----------UGUAUGUAUAUUUUUCUCCAUUCUCUGACGGAGUUUGGAUAAAAUAU ..-----........----..----------.(((((((.......(((......)----------))..............((((..(((.....)))..))))))))))). ( -13.20) >DroEre_CAF1 39238 94 + 1 CCUCUGCUGCUGCUGCAGAUUUUUUUUUGUU---UUUUAAUUUUCCCACUUUUGUA----------------UAUAUUUUUCUCCAUUUUCUGACGGAGUUUGGAUAAAAUAU ..(((((.......)))))............---......................----------------.(((((((..((((..(((.....)))..)))).))))))) ( -15.40) >consensus CC_____UGUUGAUU____UU__________UUGUUUUAAUUUUCCCACUUUUGUA__________UGUAUGUAUAUUUUUCUCCAUUCUCUGACGGAGUUUGGAUAAAAUAU .........................................................................(((((((..((((..(((.....)))..)))).))))))) (-10.86 = -10.68 + -0.19)


| Location | 18,803,388 – 18,803,492 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 76.43 |
| Mean single sequence MFE | -13.73 |
| Consensus MFE | -11.18 |
| Energy contribution | -10.92 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.81 |
| SVM decision value | 3.68 |
| SVM RNA-class probability | 0.999522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
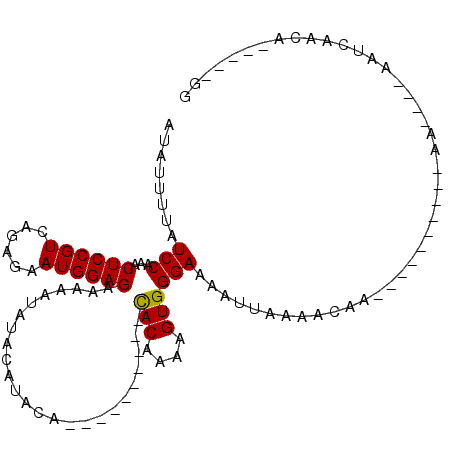
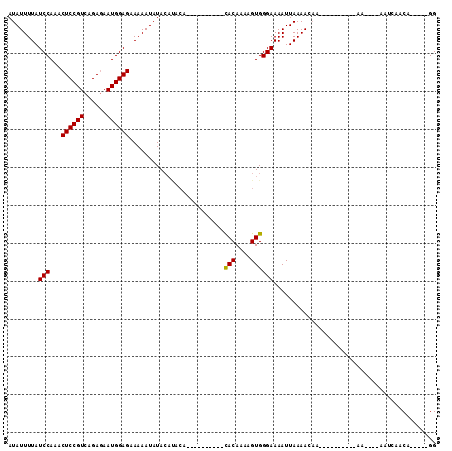
>X_DroMel_CAF1 18803388 104 - 22224390 AUAUUUUAUCCAAACUCCGUCAGAGAAUGGAGAAAAAUAUACACACAUACUCGUAUAUACAAAAGUGGGAAAAUUAAAACAAAAACAAAAAAAA----AAUCAACA-----GG ..(((((.((((..((((((......))))))....((((((..........)))))).......)))))))))....................----........-----.. ( -13.40) >DroSec_CAF1 40956 84 - 1 AUAUUUUAUCCAAACUCCGUCAGAGAAUGGAGAAAAAUAUACAUACA----------CACAAAAGUGGGAAAAUUAAAACAA----------AA----AAUCAACA-----GG ........(((...((((((......)))))).............((----------(......))))))............----------..----........-----.. ( -12.60) >DroSim_CAF1 24245 84 - 1 AUAUUUUAUCCAAACUCCGUCAGAGAAUGGAGAAAAAUAUACAUACA----------CACAAAAGUGGGAAAAUUAAAACAA----------AA----AAUCAACA-----GG ........(((...((((((......)))))).............((----------(......))))))............----------..----........-----.. ( -12.60) >DroEre_CAF1 39238 94 - 1 AUAUUUUAUCCAAACUCCGUCAGAAAAUGGAGAAAAAUAUA----------------UACAAAAGUGGGAAAAUUAAAA---AACAAAAAAAAAUCUGCAGCAGCAGCAGAGG ........(((...((((((......)))))).........----------------(((....)))))).........---............(((((.......))))).. ( -16.30) >consensus AUAUUUUAUCCAAACUCCGUCAGAGAAUGGAGAAAAAUAUACAUACA__________CACAAAAGUGGGAAAAUUAAAACAA__________AA____AAUCAACA_____GG ........(((...((((((......)))))).........................(((....))))))........................................... (-11.18 = -10.92 + -0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:19 2006