| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,801,105 – 18,801,210 |
| Length | 105 |
| Max. P | 0.931330 |

| Location | 18,801,105 – 18,801,210 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.43 |
| Mean single sequence MFE | -29.50 |
| Consensus MFE | -20.45 |
| Energy contribution | -21.64 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
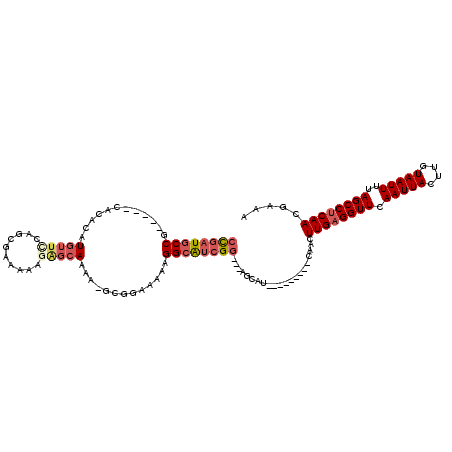
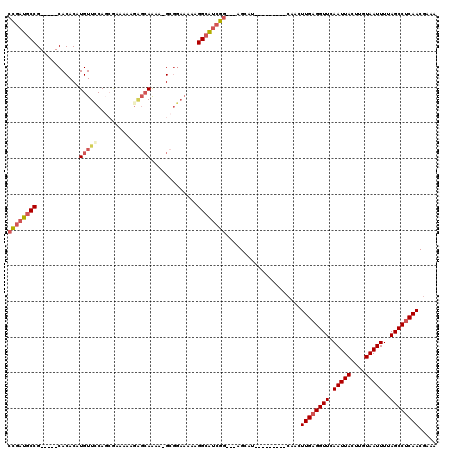
>X_DroMel_CAF1 18801105 105 + 22224390 CCGAUGCCG-----CACACAUGUUCCAGCGUAAAAGAGCAAAA-GCGGAAAAAGGCAUCGGAGGAGCAU---------CAACUUGAGGUUCAAUUACUUGUAAUUUUAGCCUCAACGAAA ((((((((.-----......((((...((........))...)-)))......))))))))........---------....((((((((.(((((....)))))..))))))))..... ( -29.52) >DroSec_CAF1 38651 103 + 1 CCGAUGCCG-----CACACAUGUUCCAGCGCACAAGAGCAAAAGGCGGAAAAAGGCAUCGG---AGCAU---------CAAGUUGAGGUUCAAUUACUUGUAAUUUUAGCCUCAACGAAA (((((((((-----((....)))(((.((((......)).....)))))....))))))))---.....---------...(((((((((.(((((....)))))..))))))))).... ( -33.50) >DroSim_CAF1 21961 95 + 1 CCGAUGCCG-----CACACAUGUUCCAGCGC--------AAAAGGCGGAAAAAGGCAUCGG---AGCAU---------CAAGUUGAGGUUCAAUUACUUGUAAUUUUAGCCUCAACGAAA (((((((((-----((....)))(((.((..--------.....)))))....))))))))---.....---------...(((((((((.(((((....)))))..))))))))).... ( -33.70) >DroEre_CAF1 36992 102 + 1 CCGAUGCCG-----CACACAUGUUCCACCAAAAAAGAGCAAAA-GCGGAAAAAGGCAUCGG---AGCAU---------CAACUUGAGGUUCAAUUACUUGUAAUUUUAGCCUCAACGAAA (((((((((-----((....)))(((..(......).((....-)))))....))))))))---.....---------....((((((((.(((((....)))))..))))))))..... ( -28.10) >DroYak_CAF1 37185 110 + 1 CCGAUGCCG-----CACACAUGUUUCAG-AAAAAAGAGCAAAA-GCGGAAAAAGGCAUCGG---AGCAACGGAGCAUCCAACUUGAGGUUCAAUUACUUGUAAUUUUAGCCUCAACGAAA (((((((((-----((....))).....-........((....-)).......))))))))---......((.....))...((((((((.(((((....)))))..))))))))..... ( -28.80) >DroPer_CAF1 50501 96 + 1 AUGAUGCCGCAGCGCACACAUGUUGCAUGG-----GGGCAGAG-G------CAGGGGCAGG---GGCAG---------CAUCUUGAGGUUCAAUUACUUGUAAUUUAAGCCGCAACAAAA ....((((...((.(...((((...)))).-----).))...)-)------))(.(((..(---(((..---------(.....)..))))(((((....)))))...))).)....... ( -23.40) >consensus CCGAUGCCG_____CACACAUGUUCCAGCGAAAAAGAGCAAAA_GCGGAAAAAGGCAUCGG___AGCAU_________CAACUUGAGGUUCAAUUACUUGUAAUUUUAGCCUCAACGAAA ((((((((............(((((..........))))).............)))))))).....................((((((((.(((((....)))))..))))))))..... (-20.45 = -21.64 + 1.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:17 2006