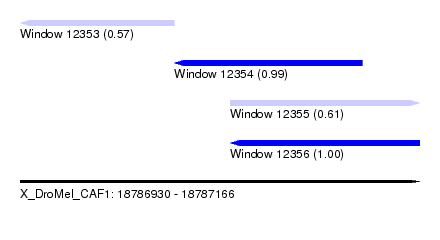
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,786,930 – 18,787,166 |
| Length | 236 |
| Max. P | 0.995879 |

| Location | 18,786,930 – 18,787,021 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 77.05 |
| Mean single sequence MFE | -18.19 |
| Consensus MFE | -13.00 |
| Energy contribution | -12.87 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568075 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18786930 91 - 22224390 AGACC------UCCAUCUC---------------CUUCUCUUCUCGGCUGUUUAGGAAGUUGUUUGCUUUCUUCAUCAGUCCAUGCACCGCACUCUUUCUAUUGCCCUGGCC (((..------........---------------...........(((((...(((((((.....)))))))....)))))..(((...))).....)))...((....)). ( -14.80) >DroSec_CAF1 33412 97 - 1 AGACC------UCCAUCUCCUC---------CUCCUUCUCUUCUCGACUGUUUAGGAAGUUGUUUGCUUUCUUCAUCAGUCCAUGGCCCACACUCCUUCUAUUGCCCUGGCC .....------...........---------..............(((((...(((((((.....)))))))....)))))...((((....................)))) ( -17.05) >DroEre_CAF1 31637 100 - 1 GGACC-----------UGCCCACCCCAUCAUCUCCU-CGCUUCUCGGCUGUUUAGGAAGUUGUUUGCUUUCCUCAUCAGUUCAUGGCCCUCACUCUUUCUAUUGCUCUGGCC ((...-----------...))...............-.(((....(((....(((((((.((...(((.....(....).....)))...))..)))))))..)))..))). ( -14.70) >DroYak_CAF1 31603 112 - 1 AGACUGAGACCCUCAGUUCCUCACCCAACAUCUCCUUCUCUUCUCGGCUGUUUAGGAAGUUGUUUGCUUUCUUCAUCAGUCCAUGGCCCUCGCUCUUUCUAUUGCUCUGGCC .(((((((...)))))))...........................(((((...(((((((.....)))))))....)))))...((((...((..........))...)))) ( -26.20) >consensus AGACC______UCCAUCUCCUC_________CUCCUUCUCUUCUCGGCUGUUUAGGAAGUUGUUUGCUUUCUUCAUCAGUCCAUGGCCCUCACUCUUUCUAUUGCCCUGGCC .............................................(((((...(((((((.....)))))))....)))))......................((....)). (-13.00 = -12.87 + -0.13)


| Location | 18,787,021 – 18,787,132 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 93.53 |
| Mean single sequence MFE | -30.71 |
| Consensus MFE | -26.16 |
| Energy contribution | -26.56 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18787021 111 - 22224390 CCAUAC-CCAGCCCCAUCUUUUUGGCUUAAUGCAGCUGCCACACGCAAGUGGGGCAUAAAAACAAAAUGUUUUGCGGGGUUCUCUAACUCGAACU------AUAACUCCCGAAUCCCG ......-...(((((.......((((...........)))).((....)))))))...(((((.....))))).((((((((.............------.........)))))))) ( -30.05) >DroSec_CAF1 33509 112 - 1 CCAUACCCCAGCCCCAUCUUUUCGGCUUAAUGCAGCUGCCACACGCAAGUGGGGCAUAAAAACAAAAUGUUUUGCGGGGUUCUCUAACUCGAACU------AUAACUCCCGAAUCCCG ..........(((((.......(((((......)))))....((....)))))))...(((((.....))))).((((((((.............------.........)))))))) ( -29.85) >DroSim_CAF1 17799 112 - 1 CCAUACCCCAGCCCCAUCUUUUUGGCUUAAUGCAGCUGCCACACGCAAGUGGGGCAUAAAAACAAAAUGUUUUGCGGGGUUCUCUAACUCGAACU------AUAACUCCCGAAUCCCG ..........(((((.......((((...........)))).((....)))))))...(((((.....))))).((((((((.............------.........)))))))) ( -30.05) >DroEre_CAF1 31737 117 - 1 CCCUGGCACAUCCCCACCUUGUUGGCUUAAUGCAGCUGCCACACGCAAGUGGGGCAUAAAAACAAAAUGUUUUGCGGGG-UCUCUAACUCGAACUACGAGUAUAACUCCCGAAUCCCG .(((.((....((((((.((((((((...........))))...))))))))))....(((((.....))))))).)))-......(((((.....)))))................. ( -33.00) >DroYak_CAF1 31715 111 - 1 CCAUGCCCCAUCCCCAUCUUUUUGGCUUAAUGCAGCUGCCACACGCAAGUGGGGCAUAAAAACAAAAUGUUUUGCGGGG-UCUCUAACUGGAACU------AUAACUCCCGAAUCCCG ..(((((((.............((((...........)))).((....))))))))).(((((.....))))).(((((-(.((.....(((...------.....))).)))))))) ( -30.60) >consensus CCAUACCCCAGCCCCAUCUUUUUGGCUUAAUGCAGCUGCCACACGCAAGUGGGGCAUAAAAACAAAAUGUUUUGCGGGGUUCUCUAACUCGAACU______AUAACUCCCGAAUCCCG .........((((((.........((.....)).((((((.(((....))).))))..(((((.....)))))))))))))..................................... (-26.16 = -26.56 + 0.40)

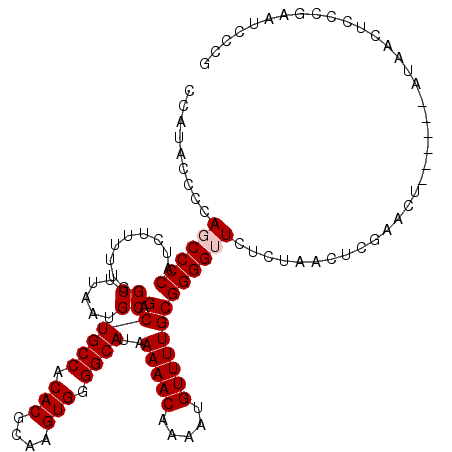
| Location | 18,787,054 – 18,787,166 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 82.07 |
| Mean single sequence MFE | -40.18 |
| Consensus MFE | -23.26 |
| Energy contribution | -24.10 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.613736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18787054 112 + 22224390 CCCCGCAAAACAUUUUGUUUUUAUGCCCCACUUGCGUGUGGCAGCUGCAUUAAGCCAAAAAGAUGGGGCUGG-GUAUGGGGCAUGGUUGGUGCUAUGGGGCGGGGGGGGG-GGG----- ((((((............(((((((((((((......))))(((((.((((..........)))).))))))-))))))))((((((....))))))..)))))).....-...----- ( -42.00) >DroSec_CAF1 33542 101 + 1 CCCCGCAAAACAUUUUGUUUUUAUGCCCCACUUGCGUGUGGCAGCUGCAUUAAGCCGAAAAGAUGGGGCUGGGGUAUGGG------------CUAUUGGGAUGGGGGGUG-GGG----- ((((((....((((..(..((((((((((((......))).(((((.((((..........)))).))))))))))))))------------....)..))))....)))-)))----- ( -38.70) >DroSim_CAF1 17832 106 + 1 CCCCGCAAAACAUUUUGUUUUUAUGCCCCACUUGCGUGUGGCAGCUGCAUUAAGCCAAAAAGAUGGGGCUGGGGUAUGGG------------CUAUUGGUGUGGGGGGUG-GGGGGUUG ((((((...(((((.....((((((((((((......))).(((((.((((..........)))).))))))))))))))------------.....))))).....)))-)))..... ( -38.90) >DroEre_CAF1 31775 101 + 1 CCCCGCAAAACAUUUUGUUUUUAUGCCCCACUUGCGUGUGGCAGCUGCAUUAAGCCAACAAGGUGGGGAUGUGCCAGGGG------------GGUUACGUACCGGGGGCG-GGG----- (((((((((((.....)))))....((((.....(.(.((((....(((((..(((.....)))...))))))))).).)------------(((.....))))))))))-)))----- ( -39.80) >DroYak_CAF1 31747 102 + 1 CCCCGCAAAACAUUUUGUUUUUAUGCCCCACUUGCGUGUGGCAGCUGCAUUAAGCCAAAAAGAUGGGGAUGGGGCAUGGG------------GCAUGAGGAUCACGAACGGGGG----- (((((((((....))))(((((((((((((..(((.....)))(((.((((...(((......))).)))).))).))))------------))))))))).......))))).----- ( -41.50) >consensus CCCCGCAAAACAUUUUGUUUUUAUGCCCCACUUGCGUGUGGCAGCUGCAUUAAGCCAAAAAGAUGGGGCUGGGGUAUGGG____________CUAUGGGGAUGGGGGGCG_GGG_____ (((((((((((.....)))))((((((((((......))))(((((.((((..........)))).))))).)))))).............................))).)))..... (-23.26 = -24.10 + 0.84)

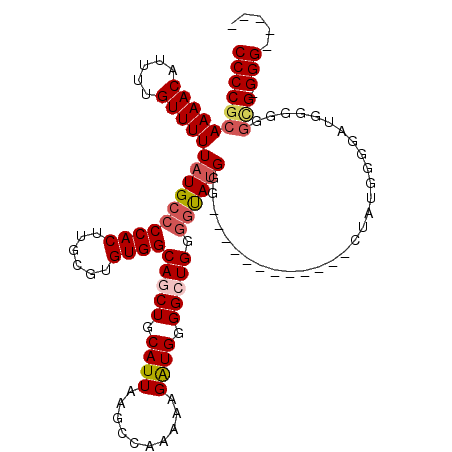
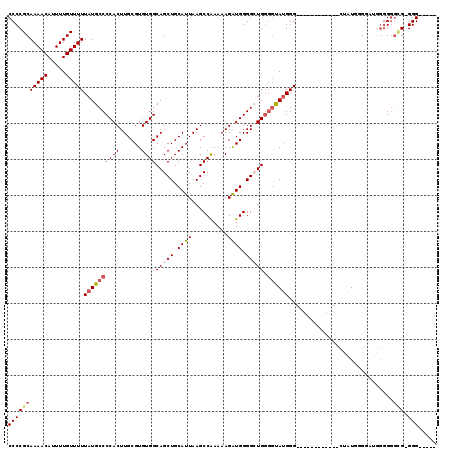
| Location | 18,787,054 – 18,787,166 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 82.07 |
| Mean single sequence MFE | -32.24 |
| Consensus MFE | -24.96 |
| Energy contribution | -24.96 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.77 |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.995879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18787054 112 - 22224390 -----CCC-CCCCCCCCCGCCCCAUAGCACCAACCAUGCCCCAUAC-CCAGCCCCAUCUUUUUGGCUUAAUGCAGCUGCCACACGCAAGUGGGGCAUAAAAACAAAAUGUUUUGCGGGG -----...-.....((((((...............(((((((....-.((((..((......))((.....)).))))....((....))))))))).(((((.....))))))))))) ( -35.80) >DroSec_CAF1 33542 101 - 1 -----CCC-CACCCCCCAUCCCAAUAG------------CCCAUACCCCAGCCCCAUCUUUUCGGCUUAAUGCAGCUGCCACACGCAAGUGGGGCAUAAAAACAAAAUGUUUUGCGGGG -----...-..................------------......((((.(((((.......(((((......)))))....((....)))))))...(((((.....)))))..)))) ( -28.20) >DroSim_CAF1 17832 106 - 1 CAACCCCC-CACCCCCCACACCAAUAG------------CCCAUACCCCAGCCCCAUCUUUUUGGCUUAAUGCAGCUGCCACACGCAAGUGGGGCAUAAAAACAAAAUGUUUUGCGGGG ........-....((((.(.......(------------(((......((((..((......))((.....)).))))...(((....)))))))...(((((.....)))))).)))) ( -29.60) >DroEre_CAF1 31775 101 - 1 -----CCC-CGCCCCCGGUACGUAACC------------CCCCUGGCACAUCCCCACCUUGUUGGCUUAAUGCAGCUGCCACACGCAAGUGGGGCAUAAAAACAAAAUGUUUUGCGGGG -----(((-(((..((((.........------------...)))).....((((((.((((((((...........))))...))))))))))....(((((.....))))))))))) ( -33.40) >DroYak_CAF1 31747 102 - 1 -----CCCCCGUUCGUGAUCCUCAUGC------------CCCAUGCCCCAUCCCCAUCUUUUUGGCUUAAUGCAGCUGCCACACGCAAGUGGGGCAUAAAAACAAAAUGUUUUGCGGGG -----.((((((.((((.....)))).------------...(((((((.............((((...........)))).((....))))))))).(((((.....))))))))))) ( -34.20) >consensus _____CCC_CACCCCCCAUCCCAAUAG____________CCCAUACCCCAGCCCCAUCUUUUUGGCUUAAUGCAGCUGCCACACGCAAGUGGGGCAUAAAAACAAAAUGUUUUGCGGGG ...................................................((((.........((.....)).((((((.(((....))).))))..(((((.....))))))))))) (-24.96 = -24.96 + 0.00)

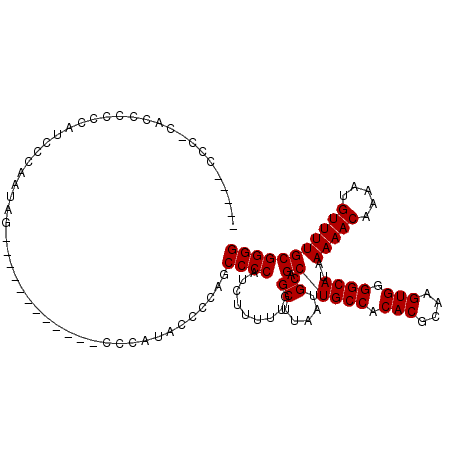
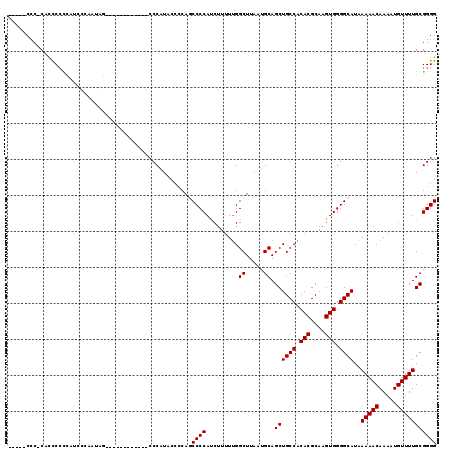
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:14 2006