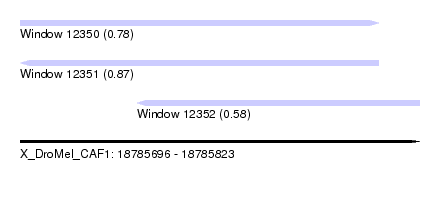
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,785,696 – 18,785,823 |
| Length | 127 |
| Max. P | 0.868939 |

| Location | 18,785,696 – 18,785,810 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 91.99 |
| Mean single sequence MFE | -32.22 |
| Consensus MFE | -25.73 |
| Energy contribution | -26.23 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18785696 114 + 22224390 AUCUGUUUUUGUAAACUGUAGGAGGAAGGUGAAAGGUGCCUGGGUGAAGUCUAAUGGUGAAAAGUGGGGACUUGGCGGCAAGUGUGUCGCAACCAGGUACCAAACAUACAGAUA-- ((((((...(((...((......)).........(((((((((...((((((...(.(....).)..)))))).((((((....))))))..)))))))))..))).)))))).-- ( -39.30) >DroSec_CAF1 32126 114 + 1 AUCUGUUUUUGUAAACUGUAGGAGGAAGGUGAAAGGUGCCUGGCUGAAGUCUAAUGGUGAAAAGUGGGAACUUCGCGGCAAGUGUGUCGCAAGCAGGUCCCAAACAUACAGAUA-- ((((((...(((...((......)).........((..((((.(((((((((.((........)).)).)))))((((((....)))))).))))))..))..))).)))))).-- ( -32.40) >DroEre_CAF1 30485 111 + 1 AUCUGUUUUUGUAAAGUGUAG---GAAGGUGAAAGGUGUCUGGGUGAAGUCUAAUGGUGAAAAGUGGGGACUUCGCGGCAAGUGUGUCGCCAGCAGGUUCCAAACAUAUAGAUA-- ((((((.((((.((..(((..---...(((((.(..((((...(((((((((...(.(....).)..)))))))))))))..)...))))).)))..)).))))...)))))).-- ( -28.50) >DroYak_CAF1 30351 113 + 1 AUCUGUUUUUGUAAACUGUAG---GAAGGUGAAAGGUGUCUGGGUGAAGUCUAAUGGUGAAAAGUGGCGACUUCGCGGCAAGUGUGUCGCAAGCAGGUUCCAAACAUAUAGAUGGA ((((((...(((...((....---..))......((..((((...((((((...((.(....).))..))))))((((((....))))))...))))..))..))).))))))... ( -28.70) >consensus AUCUGUUUUUGUAAACUGUAG___GAAGGUGAAAGGUGCCUGGGUGAAGUCUAAUGGUGAAAAGUGGGGACUUCGCGGCAAGUGUGUCGCAAGCAGGUUCCAAACAUACAGAUA__ ((((((...(((..(((..........)))....((..((((...(((((((.((........))..)))))))((((((....))))))...))))..))..))).))))))... (-25.73 = -26.23 + 0.50)

| Location | 18,785,696 – 18,785,810 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 91.99 |
| Mean single sequence MFE | -21.83 |
| Consensus MFE | -17.27 |
| Energy contribution | -17.33 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18785696 114 - 22224390 --UAUCUGUAUGUUUGGUACCUGGUUGCGACACACUUGCCGCCAAGUCCCCACUUUUCACCAUUAGACUUCACCCAGGCACCUUUCACCUUCCUCCUACAGUUUACAAAAACAGAU --.((((((.((...(((.(((((..(((.((....)).))).(((((.................)))))...))))).)))...)).............(....)....)))))) ( -21.33) >DroSec_CAF1 32126 114 - 1 --UAUCUGUAUGUUUGGGACCUGCUUGCGACACACUUGCCGCGAAGUUCCCACUUUUCACCAUUAGACUUCAGCCAGGCACCUUUCACCUUCCUCCUACAGUUUACAAAAACAGAU --.((((((.((...((..((((((.(((.((....)).)))((((((.................)))))))).))))..))...)).............(....)....)))))) ( -21.93) >DroEre_CAF1 30485 111 - 1 --UAUCUAUAUGUUUGGAACCUGCUGGCGACACACUUGCCGCGAAGUCCCCACUUUUCACCAUUAGACUUCACCCAGACACCUUUCACCUUC---CUACACUUUACAAAAACAGAU --........(((((((.....((.(((((.....)))))))((((((.................))))))..)))))))............---..................... ( -22.33) >DroYak_CAF1 30351 113 - 1 UCCAUCUAUAUGUUUGGAACCUGCUUGCGACACACUUGCCGCGAAGUCGCCACUUUUCACCAUUAGACUUCACCCAGACACCUUUCACCUUC---CUACAGUUUACAAAAACAGAU ...((((...(((((((.........(((.((....)).)))((((((.................))))))..)))))))............---.....((((....)))))))) ( -21.73) >consensus __UAUCUAUAUGUUUGGAACCUGCUUGCGACACACUUGCCGCGAAGUCCCCACUUUUCACCAUUAGACUUCACCCAGACACCUUUCACCUUC___CUACAGUUUACAAAAACAGAU ...((((...(((((((.........(((.((....)).)))((((((.................))))))..)))))))....................((((....)))))))) (-17.27 = -17.33 + 0.06)


| Location | 18,785,733 – 18,785,823 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -17.79 |
| Consensus MFE | -14.24 |
| Energy contribution | -14.05 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579106 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18785733 90 - 22224390 CCUGCCACAUCUU------UAUCUGUAUGUUUGGUACCUGGUUGCGACACACUUGCCGCCAAGUCCCCACUUUUCACCAUUAGACUUCACCCAGGC ((((.....(((.------....((...((..((.((.((((.((((.....)))).)))).))))..))....)).....))).......)))). ( -16.20) >DroSec_CAF1 32163 90 - 1 CCUGCCACAACUU------UAUCUGUAUGUUUGGGACCUGCUUGCGACACACUUGCCGCGAAGUUCCCACUUUUCACCAUUAGACUUCAGCCAGGC ((((.((((....------....))).....(((((.((..(((((.((....)).))))))).)))))....................).)))). ( -17.80) >DroEre_CAF1 30519 90 - 1 CCUGCCACAUCUC------UAUCUAUAUGUUUGGAACCUGCUGGCGACACACUUGCCGCGAAGUCCCCACUUUUCACCAUUAGACUUCACCCAGAC .............------.........((((((.....((.(((((.....)))))))((((((.................))))))..)))))) ( -19.93) >DroYak_CAF1 30385 96 - 1 CCCGCCACAUCUCCAUCUCCAUCUAUAUGUUUGGAACCUGCUUGCGACACACUUGCCGCGAAGUCGCCACUUUUCACCAUUAGACUUCACCCAGAC ............................((((((.........(((.((....)).)))((((((.................))))))..)))))) ( -17.23) >consensus CCUGCCACAUCUC______UAUCUAUAUGUUUGGAACCUGCUUGCGACACACUUGCCGCGAAGUCCCCACUUUUCACCAUUAGACUUCACCCAGAC ............................((((((.........(((.((....)).)))((((((.................))))))..)))))) (-14.24 = -14.05 + -0.19)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:11 2006