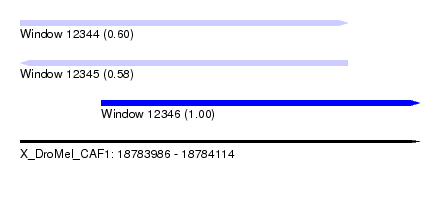
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,783,986 – 18,784,114 |
| Length | 128 |
| Max. P | 0.995899 |

| Location | 18,783,986 – 18,784,091 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.10 |
| Mean single sequence MFE | -30.04 |
| Consensus MFE | -19.42 |
| Energy contribution | -19.30 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18783986 105 + 22224390 GUUAUUACAGGU--------------CAACAUUUUUUUUUCUUGUUUAA-UAAGCCACUGGUGCGAGUUCCACCUACGCCUCUGCAAAAAAGGGAGUAGGUGUUGACUGUGCAACAACAA (((..(((((.(--------------((((..........(((((....-...(((...))))))))....((((((..((((.......)))).)))))))))))))))).)))..... ( -26.31) >DroSec_CAF1 30528 115 + 1 GUUAUUAGAGGUCGAUUAGGCGUAGUCAACAUUU---UUUCUUUUUUAU-AAACCCCCUGGUGCGAAUUCCACCUACGCCUCUGCAAA-AAGGGAGUAGGUGUUGACUGUGCAACAACAA ...................(((((((((((((((---((((((((((..-.........((((.(....)))))...((....)))))-))))))).)))))))))))))))........ ( -31.20) >DroSim_CAF1 14882 116 + 1 GUUAUUAGAGGUCGAUUAGGCGUAGUCAACAUUUU--UUUCUUUUUUAU-AAACCCCCUGGUGCGAAUUCCACCUACGCCCCUGCAAA-AAGGGAGUAGGUGUUGACUGUGCAACAACAA ...................(((((((((((.....--............-..(((....))).........((((((..((((.....-.)))).)))))))))))))))))........ ( -32.10) >DroEre_CAF1 28904 101 + 1 GUUAUUU-AGGUU----------AAUCAACGUAUU---UUAU--UUUGU-AAACCCCCUGUUGCGAGUUUCGCCUACGCCUCUGUAA--AAGGGAGUAGGUGUUGACUGUGCAACAACAA .......-.((((----------...(((.(((..---.)))--.))).-.))))...(((((((((((.(((((((..((((....--.)))).)))))))..)))).))))))).... ( -30.20) >DroYak_CAF1 28778 115 + 1 GUUAUUAGAGGUCGAUUGUGCAGAAUACACAUCUU---UUAU--UUUAUUAAGCCCCCUGUGGCGAAUUGCACCUACGCCUCUGCAAAAAAGGGAGUAGGUGUUGACUGUGCAACAACAA ..........((.(.(((..(((............---....--........(((......))).....((((((((..((((.......)))).))))))))...)))..)))).)).. ( -30.40) >consensus GUUAUUAGAGGUCGAUU__GC__AAUCAACAUUUU__UUUCUU_UUUAU_AAACCCCCUGGUGCGAAUUCCACCUACGCCUCUGCAAA_AAGGGAGUAGGUGUUGACUGUGCAACAACAA ..........................................................((.((((..(..(((((((..((((.......)))).)))))))..)....)))).)).... (-19.42 = -19.30 + -0.12)



| Location | 18,783,986 – 18,784,091 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.10 |
| Mean single sequence MFE | -28.60 |
| Consensus MFE | -16.95 |
| Energy contribution | -17.55 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18783986 105 - 22224390 UUGUUGUUGCACAGUCAACACCUACUCCCUUUUUUGCAGAGGCGUAGGUGGAACUCGCACCAGUGGCUUA-UUAAACAAGAAAAAAAAAUGUUG--------------ACCUGUAAUAAC ..(((((((((..((((((((((((..(((((.....))))).)))))).....((((....))))....-...................))))--------------)).))))))))) ( -28.80) >DroSec_CAF1 30528 115 - 1 UUGUUGUUGCACAGUCAACACCUACUCCCUU-UUUGCAGAGGCGUAGGUGGAAUUCGCACCAGGGGGUUU-AUAAAAAAGAAA---AAAUGUUGACUACGCCUAAUCGACCUCUAAUAAC ..((((..((..(((((((((((((..((((-......)))).)))))).((((((.(....).))))))-............---....)))))))..)).....)))).......... ( -28.10) >DroSim_CAF1 14882 116 - 1 UUGUUGUUGCACAGUCAACACCUACUCCCUU-UUUGCAGGGGCGUAGGUGGAAUUCGCACCAGGGGGUUU-AUAAAAAAGAAA--AAAAUGUUGACUACGCCUAAUCGACCUCUAAUAAC ..((((..((..(((((((((((((.(((((-.....))))).)))))).((((((.(....).))))))-............--.....)))))))..)).....)))).......... ( -31.90) >DroEre_CAF1 28904 101 - 1 UUGUUGUUGCACAGUCAACACCUACUCCCUU--UUACAGAGGCGUAGGCGAAACUCGCAACAGGGGGUUU-ACAAA--AUAA---AAUACGUUGAUU----------AACCU-AAAUAAC ..((((((....(((((((.(((((..((((--.....)))).)))))..((((((.(....).))))))-.....--....---.....)))))))----------.....-.)))))) ( -25.00) >DroYak_CAF1 28778 115 - 1 UUGUUGUUGCACAGUCAACACCUACUCCCUUUUUUGCAGAGGCGUAGGUGCAAUUCGCCACAGGGGGCUUAAUAAA--AUAA---AAGAUGUGUAUUCUGCACAAUCGACCUCUAAUAAC ..(((((((....(((..(((((((..(((((.....))))).)))))))......(((......)))........--....---..(((((((.....)))).))))))...))))))) ( -29.20) >consensus UUGUUGUUGCACAGUCAACACCUACUCCCUU_UUUGCAGAGGCGUAGGUGGAAUUCGCACCAGGGGGUUU_AUAAA_AAGAAA__AAAAUGUUGACU__GC__AAUCGACCUCUAAUAAC ..((((((((..(((...(((((((..((((.......)))).)))))))..))).))...(((((...........................................))))))))))) (-16.95 = -17.55 + 0.60)


| Location | 18,784,012 – 18,784,114 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 90.87 |
| Mean single sequence MFE | -28.38 |
| Consensus MFE | -23.76 |
| Energy contribution | -24.04 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.995899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18784012 102 + 22224390 CUUGUUUAA-UAAGCCACUGGUGCGAGUUCCACCUACGCCUCUGCAAAAAAGGGAGUAGGUGUUGACUGUGCAACAACAAAGUUGGCAAUAAAAAUGCAACCA .........-...((((((.(((((((((.(((((((..((((.......)))).)))))))..)))).))).)).....)).))))................ ( -27.90) >DroSec_CAF1 30565 101 + 1 CUUUUUUAU-AAACCCCCUGGUGCGAAUUCCACCUACGCCUCUGCAAA-AAGGGAGUAGGUGUUGACUGUGCAACAACAAAGUUGGCAAUAAAAAUGCAACCA .........-.........((((((......((((((..((((.....-.)))).))))))((..(((.((......)).)))..))........))).))). ( -26.50) >DroSim_CAF1 14920 101 + 1 CUUUUUUAU-AAACCCCCUGGUGCGAAUUCCACCUACGCCCCUGCAAA-AAGGGAGUAGGUGUUGACUGUGCAACAACAAAGUUGGCAAUAAAAAUGCAACCA .........-.........((((((......((((((..((((.....-.)))).))))))((..(((.((......)).)))..))........))).))). ( -29.50) >DroEre_CAF1 28930 98 + 1 AU--UUUGU-AAACCCCCUGUUGCGAGUUUCGCCUACGCCUCUGUAA--AAGGGAGUAGGUGUUGACUGUGCAACAACAAAGUUGGCAAUAAAAAUGCAACCA ..--.((((-.(((....(((((((((((.(((((((..((((....--.)))).)))))))..)))).))))))).....))).)))).............. ( -30.50) >DroYak_CAF1 28815 101 + 1 AU--UUUAUUAAGCCCCCUGUGGCGAAUUGCACCUACGCCUCUGCAAAAAAGGGAGUAGGUGUUGACUGUGCAACAACAAAGUUGGCAAUAAAAAUGCAACCA ..--........(((......)))...((((((((((..((((.......)))).)))))(((..(((.((......)).)))..))).......)))))... ( -27.50) >consensus CUU_UUUAU_AAACCCCCUGGUGCGAAUUCCACCUACGCCUCUGCAAA_AAGGGAGUAGGUGUUGACUGUGCAACAACAAAGUUGGCAAUAAAAAUGCAACCA ...................((((((......((((((..((((.......)))).))))))((..(((.((......)).)))..))........))).))). (-23.76 = -24.04 + 0.28)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:05 2006