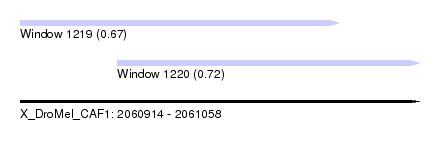
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,060,914 – 2,061,058 |
| Length | 144 |
| Max. P | 0.720914 |

| Location | 2,060,914 – 2,061,029 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.72 |
| Mean single sequence MFE | -45.28 |
| Consensus MFE | -25.72 |
| Energy contribution | -27.17 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670327 |
| Prediction | RNA |
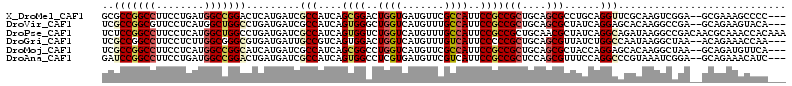
Download alignment: ClustalW | MAF
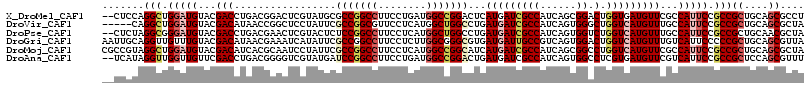
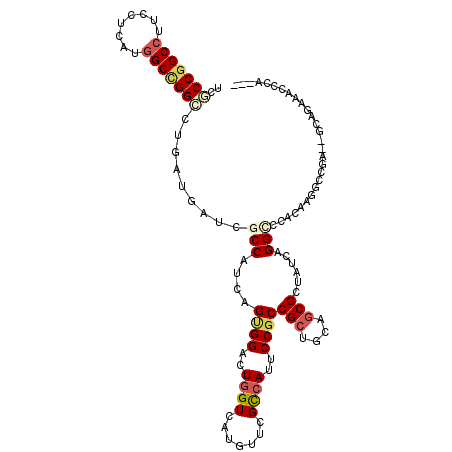
>X_DroMel_CAF1 2060914 115 + 22224390 --CUCCAGGCUGGAUGUACGACCUGACGGACUCGUAUGCGCCGGCCUUCCUGAUGGCCGGACUCAUGAUCGCCAUCAGCGGACUGGUGAUGUUCGCCAUUCCGCCGCUGCAGCGCCU --....(((((((..((((((((....))..))))))...)))))))...(((((((.((.(....).)))))))))(((((.(((((.....))))).)))))(((....)))... ( -48.50) >DroVir_CAF1 3140 112 + 1 -----CAGGCUGGAUGUACGACAUAACCGGCUCCUAUUCGCCGGCGUUCCUCAUGGCUGGCCUGAUGAUCGCCAUCAGUGGGCUGGUCAUGUUUGCCAUUCCGCCGCUGCAGCGCUA -----..(((.(((((...(((((.((((((((......((((((..........))))))((((((.....)))))).)))))))).)))))...))))).)))((....)).... ( -46.60) >DroPse_CAF1 3345 115 + 1 --CUCUAGGCGGGAUGUACGACCUGACGAACUCGUACUCUCCGGCCUUCCUCAUGGCUGGCCUGAUGAUCGCCAUCAGUGGUCUGGUCAUGUUUGCCAUUCCGCCGCUGCAACGCUA --.....(((((((((.((((((.(((.............((((((........))))))(((((((.....)))))).)))).))))..))....)))))))))((......)).. ( -43.60) >DroGri_CAF1 2618 117 + 1 AAUUGCAGGUUGUUUGUACGACAUAACGAAAUCAUAUUCGCCGGCCUUCCUCUUGGCGGGCGUGAUGAUUGCCGUCAGUGGACUGGUCAUGUUUGUCAUUCCCCCGCUGCAGCGUUA ........(((((....))))).((((..((((((...((((.(((........))).))))..))))))((.(.((((((...((..(((.....)))..))))))))).)))))) ( -40.10) >DroMoj_CAF1 3328 117 + 1 CGCCGUAGGCUGGAUGUACGACAUCACGCAAUCCUAUUCGCCGGCCUUCCUCAUGGCCGGCAUCAUGAUCGCCAUCAGCGGCCUGGUCAUGUUCGCCAUUCCGCCGCUGCAGCGCUA (((....(((.((((...((......))..)))).....(((((((........))))))).........)))..(((((((.((((.......))))....)))))))..)))... ( -45.10) >DroAna_CAF1 1965 115 + 1 --UCAUAGGUUGGUUGUUCGACCUGACGGGGUCGUAUGAUCCGGCCUUCCUGAUGGCCGGACUGAUGAUCGCCAUCAGUGGCCUCGUGAUGUUCGUCAUUCCGCCGCUCCAGCGUUU --...((((((((....))))))))((((((((.......((((((........))))))(((((((.....)))))))))))))))(((....))).......(((....)))... ( -47.80) >consensus __CUCCAGGCUGGAUGUACGACAUGACGGACUCGUAUUCGCCGGCCUUCCUCAUGGCCGGCCUGAUGAUCGCCAUCAGUGGACUGGUCAUGUUCGCCAUUCCGCCGCUGCAGCGCUA .......(((.(((((...(((.................(((((((........)))))))...(((((((((......)).).)))))))))...))))).)))((....)).... (-25.72 = -27.17 + 1.45)
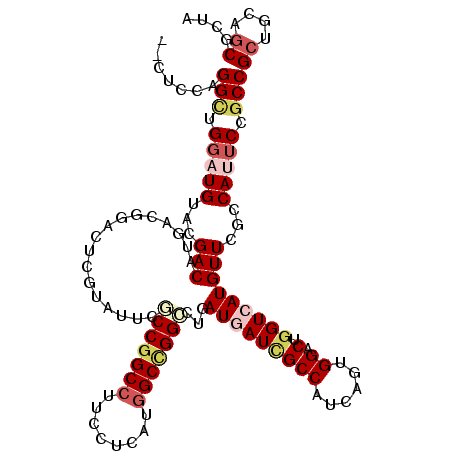
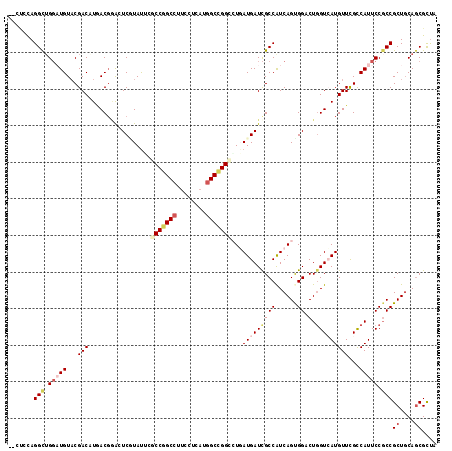
| Location | 2,060,949 – 2,061,058 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.47 |
| Mean single sequence MFE | -42.67 |
| Consensus MFE | -23.42 |
| Energy contribution | -24.12 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.720914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2060949 109 + 22224390 GCGCCGGCCUUCCUGAUGGCCGGACUCAUGAUCGCCAUCAGCGGACUGGUGAUGUUCGCCAUUCCGCCGCUGCAGCGCCUGCAGGUUCGCAAGUCGGA--GCGAAAGCCCC--- (((.((((....((((((((.((.(....).)))))))))).(((.(((((.....))))).)))))))((((((...))))))...))).....((.--((....)))).--- ( -48.60) >DroVir_CAF1 3172 109 + 1 UCGCCGGCGUUCCUCAUGGCUGGCCUGAUGAUCGCCAUCAGUGGGCUGGUCAUGUUUGCCAUUCCGCCGCUGCAGCGCUAUCAGGAGCACAAGGCCGA--GCAGAAGUACA--- ..((((((((((((.(((((.(((((((((.....)))))).((..(((.((....)))))..)))))((....))))))).)))))).....)))).--)).........--- ( -44.20) >DroPse_CAF1 3380 114 + 1 UCUCCGGCCUUCCUCAUGGCUGGCCUGAUGAUCGCCAUCAGUGGUCUGGUCAUGUUUGCCAUUCCGCCGCUGCAACGCUAUCAGGCAGAUAAGGCCGACAACGCAAACCACAAA .....((((..((....))..))))(((((.....)))))(((((.((((..((((.(((..((.(((...((...)).....))).))...))).)))))).)).)))))... ( -38.70) >DroGri_CAF1 2655 109 + 1 UCGCCGGCCUUCCUCUUGGCGGGCGUGAUGAUUGCCGUCAGUGGACUGGUCAUGUUUGUCAUUCCCCCGCUGCAGCGUUAUCUGGCCAAUAAGGCUAA--ACAGAAACCAA--- .((((.(((........))).)))).((((((.((.(.((((((...((..(((.....)))..))))))))).))))))))(((((.....))))).--...........--- ( -38.70) >DroMoj_CAF1 3365 109 + 1 UCGCCGGCCUUCCUCAUGGCCGGCAUCAUGAUCGCCAUCAGCGGCCUGGUCAUGUUCGCCAUUCCGCCGCUGCAGCGCUACCAGGAGCACAAGGCUAA--GCAGAUGUUCA--- ..(((((((........)))))))(((.((...(((..(((((((.((((.......))))....)))))))..(.(((......))).)..)))...--.))))).....--- ( -42.60) >DroAna_CAF1 2000 109 + 1 GAUCCGGCCUUCCUGAUGGCCGGACUGAUGAUCGCCAUCAGUGGCCUCGUGAUGUUCGUCAUUCCGCCGCUCCAGCGUUUCCAGGCCCGUAAAUCGGA--GCAGAAACAUC--- ...((((((........))))))(((((((.....)))))))(((...(((((....)))))...)))((....))(((((...(((((.....))).--)).)))))...--- ( -43.20) >consensus UCGCCGGCCUUCCUCAUGGCCGGCCUGAUGAUCGCCAUCAGUGGACUGGUCAUGUUCGCCAUUCCGCCGCUGCAGCGCUAUCAGGCCCACAAGGCCGA__GCAGAAACCCA___ ..(((((((........))))))).........(((....((((..((((.......))))..))))(((....)))......)))............................ (-23.42 = -24.12 + 0.69)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:57 2006