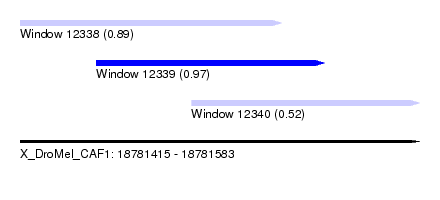
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,781,415 – 18,781,583 |
| Length | 168 |
| Max. P | 0.973046 |

| Location | 18,781,415 – 18,781,525 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -25.98 |
| Energy contribution | -25.91 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18781415 110 + 22224390 AAAGGAGGACGCAAAAGGAACAAGUCCCAGGCAGAACCUUUAACGUUCUAUCGACAAAAGUCUCAUUGUCUCACGGGCCACAUGGCCCUGUAAUCAAAUAUCAAUGAGAU ...((((((((..(((((.....((.....))....)))))..)))))).)).......(((((((((......(((((....)))))((....)).....))))))))) ( -31.20) >DroSec_CAF1 27900 110 + 1 AAAGGAGGACGCAAAAGGAACAAGUCCCAGGCAGAACCUUUAACGGCCUACCGACAAAAGUCUCAUUGUCUCACGGGCCACAUGGCCCUGUAAUCAAAUGUCAAUGAGAU ...(((((.((..(((((.....((.....))....)))))..)).))).)).......(((((((((.(....(((((....)))))((....))...).))))))))) ( -33.20) >DroEre_CAF1 26391 110 + 1 AAAGGAGGGCGGAAAAGGAACAAGUCCCAGGCAGAACCUUUAACGGUCUAUCGACAAAAGUCUCAUUGUCUCACGGGCCACAUGGCCCUGCAAUCAAAUAUCAAUGAGAU ...(((((.((..(((((.....((.....))....)))))..)).))).)).......(((((((((...((.(((((....)))))))...........))))))))) ( -29.74) >DroYak_CAF1 26081 110 + 1 AAAGGCGGGCGGGAAAGGAACAAGUUUCAGGCAGAAUCUUUAACGGUCUACCGACAAAAAUCUCAUUGUCUCACGGAGCUCGUGGCCCUGCAAUCAAAUAUCAAUGAGAU ...((..(((.(..(((((....((.....))....)))))..).)))..)).......(((((((((...((((.....))))((...))..........))))))))) ( -25.80) >consensus AAAGGAGGACGCAAAAGGAACAAGUCCCAGGCAGAACCUUUAACGGUCUACCGACAAAAGUCUCAUUGUCUCACGGGCCACAUGGCCCUGCAAUCAAAUAUCAAUGAGAU ...(((((.((..(((((.....((.....))....)))))..)).))).)).......(((((((((...((.(((((....)))))))...........))))))))) (-25.98 = -25.91 + -0.06)



| Location | 18,781,447 – 18,781,543 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 90.97 |
| Mean single sequence MFE | -26.80 |
| Consensus MFE | -22.74 |
| Energy contribution | -22.74 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.973046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18781447 96 + 22224390 AGAACCUUUAACGUUCUAUCGACAAAAGUCUCAUUGUCUCACGGGCCACAUGGCCCUGUAAUCAAAUAUCAAUGAGAUGAAGGAGGUACUAUCAUU ((.((((((..((......))......(((((((((......(((((....)))))((....)).....)))))))))...)))))).))...... ( -27.10) >DroSec_CAF1 27932 96 + 1 AGAACCUUUAACGGCCUACCGACAAAAGUCUCAUUGUCUCACGGGCCACAUGGCCCUGUAAUCAAAUGUCAAUGAGAUGAAGGACGUACUAUCAUU ....(((((..(((....)))......(((((((((.(....(((((....)))))((....))...).))))))))))))))............. ( -29.50) >DroEre_CAF1 26423 96 + 1 AGAACCUUUAACGGUCUAUCGACAAAAGUCUCAUUGUCUCACGGGCCACAUGGCCCUGCAAUCAAAUAUCAAUGAGAUGGAGGCGGUACUAUCAUC ((.(((.......(((....)))....(((((...((((((.(((((....)))))((....))........)))))).)))))))).))...... ( -28.30) >DroYak_CAF1 26113 96 + 1 AGAAUCUUUAACGGUCUACCGACAAAAAUCUCAUUGUCUCACGGAGCUCGUGGCCCUGCAAUCAAAUAUCAAUGAGAUGGAGGAGGCACUAUCAUC .(..(((((..(((....)))......(((((((((...((((.....))))((...))..........))))))))).)))))..)......... ( -22.30) >consensus AGAACCUUUAACGGUCUACCGACAAAAGUCUCAUUGUCUCACGGGCCACAUGGCCCUGCAAUCAAAUAUCAAUGAGAUGAAGGAGGUACUAUCAUC ....(((((..(((....)))......(((((((((...((.(((((....)))))))...........))))))))))))))............. (-22.74 = -22.74 + 0.00)

| Location | 18,781,487 – 18,781,583 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 88.02 |
| Mean single sequence MFE | -19.75 |
| Consensus MFE | -14.95 |
| Energy contribution | -14.70 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18781487 96 + 22224390 ACGGGCCACAUGGCCCUGUAAUCAAAUAUCAAUGAGAUGAAGGAGGUACUAUCAUUCUGCACACAGAAAAAUCGUUGUAAUUAACAUAAUUAACAC ..(((((....)))))(((..........((((((..(((((......)).)))(((((....)))))...))))))((((((...))))))))). ( -20.70) >DroSec_CAF1 27972 96 + 1 ACGGGCCACAUGGCCCUGUAAUCAAAUGUCAAUGAGAUGAAGGACGUACUAUCAUUCUGCACACAGAAAAAUCGAUGUAAUUAACAUAGUUAACAC ..(((((....)))))..........(((.(((......((..((((.......(((((....)))))......))))..))......))).))). ( -19.12) >DroEre_CAF1 26463 96 + 1 ACGGGCCACAUGGCCCUGCAAUCAAAUAUCAAUGAGAUGGAGGCGGUACUAUCAUCCUGCACCCGGAACAAUUGAUCAAAUUAACAUAAUUAACAU ..(((((....)))))...........((((((....(((..((((..........))))..))).....)))))).................... ( -23.40) >DroYak_CAF1 26153 96 + 1 ACGGAGCUCGUGGCCCUGCAAUCAAAUAUCAAUGAGAUGGAGGAGGCACUAUCAUCCUGCACACGAAAAAAUUGAUCUAAUUAACAUAAUUAACAC ..(((....(((.((((.((.(((........)))..)).))).).))).....)))....................................... ( -15.80) >consensus ACGGGCCACAUGGCCCUGCAAUCAAAUAUCAAUGAGAUGAAGGAGGUACUAUCAUCCUGCACACAGAAAAAUCGAUCUAAUUAACAUAAUUAACAC ..(((((....)))))((((.(((........)))(((((((......)).))))).))))................................... (-14.95 = -14.70 + -0.25)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:00 2006