| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,779,644 – 18,779,843 |
| Length | 199 |
| Max. P | 0.973606 |

| Location | 18,779,644 – 18,779,739 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 76.77 |
| Mean single sequence MFE | -17.76 |
| Consensus MFE | -14.92 |
| Energy contribution | -14.70 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18779644 95 + 22224390 CGCUCCCGCUCCCGCUCCCAAUUCCAAUCCCGUU------------CCUGUUCCCACAACUUCCCUUUUAACCGGGAGUGCCUUUUGUGACUGGGUACUCUCAAUCA (((((((((....))...................------------..(((....)))...............))))))).......(((.((((....)))).))) ( -16.80) >DroSec_CAF1 26135 83 + 1 CGC------------UCCCAAUCCCAAUCCCGUU------------CCUGUUCCCACAACUUCCCUUUUACCCGGGAGUGCCUUUUGUGACUGGGUACUUUCAAUCA .(.------------.((((.((.(((...((((------------((((......................))))))))....))).)).))))..)......... ( -14.05) >DroEre_CAF1 24632 95 + 1 CGU------------UCCCGUUCCCGUUCCCAUUCCCGCUAGCAAUCCCGUUCCCACAACUUCCCUUUUAGCCGGGAGUGCCUUUUGUGACUGGGUACUCUCAAUCA .((------------.((((.((.((....((((((((((((.......(((.....))).......)))).)))))))).....)).)).)))).))......... ( -22.44) >consensus CGC____________UCCCAAUCCCAAUCCCGUU____________CCUGUUCCCACAACUUCCCUUUUAACCGGGAGUGCCUUUUGUGACUGGGUACUCUCAAUCA .........................................................................((((((((((.........))))))))))..... (-14.92 = -14.70 + -0.22)

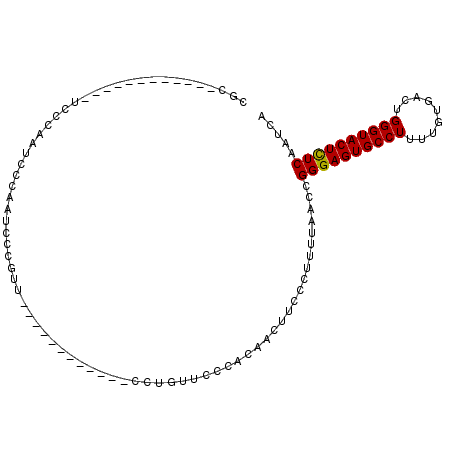

| Location | 18,779,644 – 18,779,739 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 76.77 |
| Mean single sequence MFE | -25.07 |
| Consensus MFE | -16.76 |
| Energy contribution | -16.32 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18779644 95 - 22224390 UGAUUGAGAGUACCCAGUCACAAAAGGCACUCCCGGUUAAAAGGGAAGUUGUGGGAACAGG------------AACGGGAUUGGAAUUGGGAGCGGGAGCGGGAGCG ............(((.(((......))).(((((((((..((......((.((....)).)------------)......))..))))))))).))).((....)). ( -27.60) >DroSec_CAF1 26135 83 - 1 UGAUUGAAAGUACCCAGUCACAAAAGGCACUCCCGGGUAAAAGGGAAGUUGUGGGAACAGG------------AACGGGAUUGGGAUUGGGA------------GCG .........((.(((((((.(((.......((((........))))..((.((....)).)------------)......))).))))))).------------)). ( -23.60) >DroEre_CAF1 24632 95 - 1 UGAUUGAGAGUACCCAGUCACAAAAGGCACUCCCGGCUAAAAGGGAAGUUGUGGGAACGGGAUUGCUAGCGGGAAUGGGAACGGGAACGGGA------------ACG .........((.((((.((.(....((((.(((((.(((.((......)).)))...))))).))))....))).)))).)).....((...------------.)) ( -24.00) >consensus UGAUUGAGAGUACCCAGUCACAAAAGGCACUCCCGGCUAAAAGGGAAGUUGUGGGAACAGG____________AACGGGAUUGGGAUUGGGA____________GCG ............(((.(((......)))..(((((..............(.((....)).)..............)))))........)))................ (-16.76 = -16.32 + -0.44)

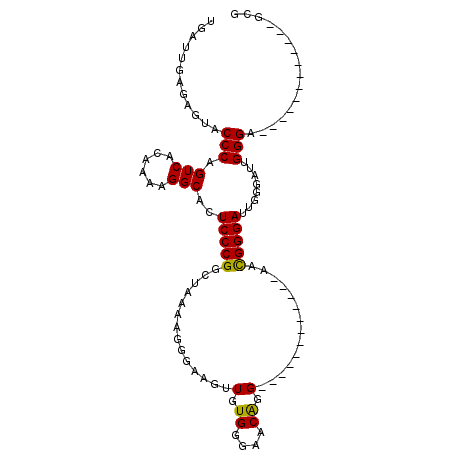

| Location | 18,779,678 – 18,779,774 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 82.98 |
| Mean single sequence MFE | -32.65 |
| Consensus MFE | -27.31 |
| Energy contribution | -29.62 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.84 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18779678 96 + 22224390 ------------------CCUGUUCCCACAACUUCCCUUUUAACCGGGAGUGCCUUUUGUGACUGGGUACUCUCAAUCAAAGCCGGCCC-----GGGCCGGGACUUUGGACUCGAGCUC ------------------...((((....................((((((((((.........))))))))))..(((((((((((..-----..)))))..))))))....)))).. ( -33.80) >DroSec_CAF1 26157 101 + 1 ------------------CCUGUUCCCACAACUUCCCUUUUACCCGGGAGUGCCUUUUGUGACUGGGUACUUUCAAUCAAAGCCGGCCCGGACCGGGCCGGGACUUUGGACUCGAGCUC ------------------...((((....................(..(((((((.........)))))))..)..((((((((((((((...))))))))..))))))....)))).. ( -37.30) >DroEre_CAF1 24660 97 + 1 ------------AGCAAUCCCGUUCCCACAACUUCCCUUUUAGCCGGGAGUGCCUUUUGUGACUGGGUACUCUCAAUCAAAGCC----------GCGCCGGGACUUUGGACUCGAGCUC ------------.........((((....................((((((((((.........))))))))))..((((((((----------......)).))))))....)))).. ( -26.30) >DroYak_CAF1 24361 109 + 1 ACCGCUACUGCUACCAAUCCCGUUCCCACAACUUCCCUUUUAGCCGGGAGUGCCUUUUGUGACUGGGUACUCUCAAUCAAAGCC----------GGGCCGGGACUUUGGACUCGAGCUC ...(((.......(((((((((..(((....((........))..((((((((((.........))))))))))..........----------))).)))))..)))).....))).. ( -33.20) >consensus __________________CCCGUUCCCACAACUUCCCUUUUAGCCGGGAGUGCCUUUUGUGACUGGGUACUCUCAAUCAAAGCC__________GGGCCGGGACUUUGGACUCGAGCUC .....................((((....................((((((((((.........))))))))))..(((((((((((((.....)))))))..))))))....)))).. (-27.31 = -29.62 + 2.31)

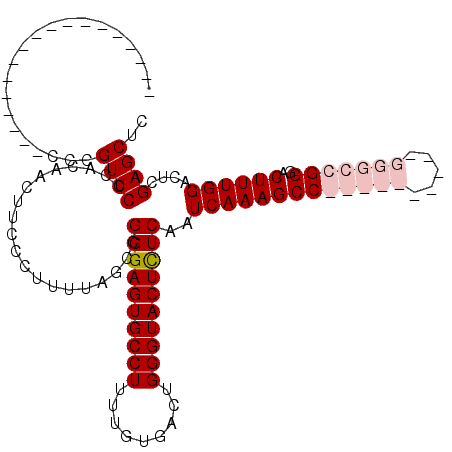

| Location | 18,779,739 – 18,779,843 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 91.04 |
| Mean single sequence MFE | -28.71 |
| Consensus MFE | -22.85 |
| Energy contribution | -25.35 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.919032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18779739 104 - 22224390 CUCCCCCAUUUUGGUAGUUUCUGAAACUCCCCUUCGUUUUUUUC-CCAUUUUGUUCCCUGCCAAAAAGGGGAGCUCGAGUCCAAAGUCCCGGCCC-----GGGCCGGCUU ((((((..(((((((((.....(((((........)))))....-............))))))))).))))))...............(((((..-----..)))))... ( -30.80) >DroSec_CAF1 26218 110 - 1 CUCCCCCAUUUUGGUAGUUUCUGAAACUCCCCUUCAUUUUUUUCUCCAUUCUGCUCCCUGCCAAAAAGGGGAGCUCGAGUCCAAAGUCCCGGCCCGGUCCGGGCCGGCUU .......(((((((.......((((.......))))...........((((.(((((((........)))))))..))))))))))).((((((((...))))))))... ( -36.20) >DroEre_CAF1 24727 98 - 1 CUCCCCCAUUUUGGUAGUUUCUGAAACUCCCCUUCAUUUUUU-C-CCAUUUUGCUCCCUGCCAAAAAGGGGAGCUCGAGUCCAAAGUCCCGGCGC----------GGCUU ...((((.(((((((((....((((.......))))......-.-.(.....)....))))))))).))))(((.((.(.((........))).)----------)))). ( -23.60) >DroYak_CAF1 24440 98 - 1 CUCCCCCAUUUUGGUAGUUUCUGAAACUCCCCUUCAUUUUUU-C-CCAUUUUUCUCCCUGCCAAAAAGGGGAGCUCGAGUCCAAAGUCCCGGCCC----------GGCUU ...((((.(((((((((.....((((................-.-.....))))...))))))))).))))(((.((.(.((........))).)----------)))). ( -24.25) >consensus CUCCCCCAUUUUGGUAGUUUCUGAAACUCCCCUUCAUUUUUU_C_CCAUUUUGCUCCCUGCCAAAAAGGGGAGCUCGAGUCCAAAGUCCCGGCCC__________GGCUU ((((((..(((((((((........................................))))))))).))))))...............(((((((.....)))))))... (-22.85 = -25.35 + 2.50)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:57 2006