
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,779,346 – 18,779,535 |
| Length | 189 |
| Max. P | 0.751182 |

| Location | 18,779,346 – 18,779,457 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 80.59 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -21.42 |
| Energy contribution | -22.55 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.533955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18779346 111 - 22224390 UUGUGUCUUAGCCUGGCCAAAAACUGGCAUGCCAAAAAGG----GGGAGAAAUCAGUGAAAGGCUUGACUCGGAAACGGGGACAUUCUCCAGACUUUAAGUCGCAAAGACCAGCA (((.(((((.((...((((.....)))).......((((.----.((((((.(((((.....)).)))((((....))))....))))))...)))).....)).)))))))).. ( -33.30) >DroSec_CAF1 25840 110 - 1 UUGUGUCUUAGCCUGGCCAAAAGCUGGCAUGCCAAAAUAG----G-GGGAAAUCAGUGAAAGGCUUGACGUGGAAACAGGGACAUUCUCCAGGCUUUAAGUCGCAAAUACCCGCA (((((.((((((((((((....(((((..(.((.......----.-)).)..)))))....)))....(.((....)).).........))))))..))).)))))......... ( -33.60) >DroEre_CAF1 24353 98 - 1 UUGUGUCUUUGCCUGGCCAAAAACUGGCAUACCCAAAUGGGCAGG-AGGAAAUCAGUGAAAGGCUUGAC----------------UCUUCAGGCUUUAAGUCGCAAAUCUAGGCA ...((((((..((((((((.....))))...(((....)))))))-..)).....(((((((((((((.----------------...)))))))))...)))).......)))) ( -30.90) >DroYak_CAF1 24035 110 - 1 UUGUGUCUUGGCCUGGCCAAAAACUGGCAUACCAAAAUGG----G-GGGAAAUCAGUGAAAGGCUUGACUCGGAAACAGGCACACUCUCCAAGCUUUAAAUCGCAAAUCCAGGCA ..........(((((((((.....))))...((.....))----.-.........((((((((((((....(....)((.....))...))))))))...)))).....))))). ( -29.40) >consensus UUGUGUCUUAGCCUGGCCAAAAACUGGCAUACCAAAAUGG____G_GGGAAAUCAGUGAAAGGCUUGACUCGGAAACAGGGACAUUCUCCAGGCUUUAAGUCGCAAAUACAGGCA (((((.(((((((((((((.....))))..................((((..(((((.....)).)))((((....)))).....))))))))))..))).)))))......... (-21.42 = -22.55 + 1.13)


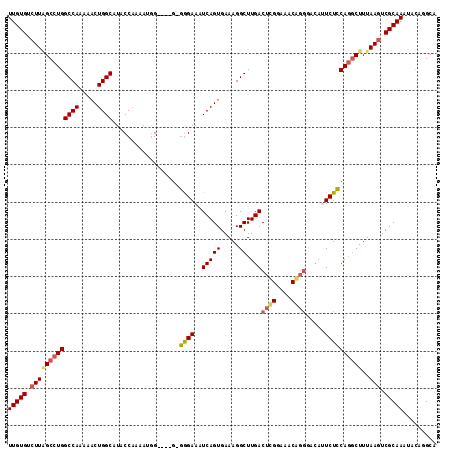
| Location | 18,779,381 – 18,779,497 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.52 |
| Mean single sequence MFE | -33.30 |
| Consensus MFE | -23.87 |
| Energy contribution | -24.56 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564424 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18779381 116 - 22224390 AAGGGCUCCGCAGCCCCAACCCUGAAAUGAAAUACAGUCAUUGUGUCUUAGCCUGGCCAAAAACUGGCAUGCCAAAAAGG----GGGAGAAAUCAGUGAAAGGCUUGACUCGGAAACGGG ..(((((....)))))(((.(((..............((((((..((((..(((.((((.....)))).........)))----..))))...)))))).))).))).((((....)))) ( -35.06) >DroSec_CAF1 25875 115 - 1 AAGGGCUCCGCAGCCCCAACCCUGAAAUGAAACACAGUCAUUGUGUCUUAGCCUGGCCAAAAGCUGGCAUGCCAAAAUAG----G-GGGAAAUCAGUGAAAGGCUUGACGUGGAAACAGG ..(((((....)))))....((((...(((.......)))(..((((..(((((.((((.....)))).(.((......)----)-.)............))))).))))..)...)))) ( -32.00) >DroEre_CAF1 24383 108 - 1 AAGGGCUCCACAGCCCCAACCCUGAAAUGAAACACAGUCAUUGUGUCUUUGCCUGGCCAAAAACUGGCAUACCCAAAUGGGCAGG-AGGAAAUCAGUGAAAGGCUUGAC----------- ..(((((....)))))(((.(((..............((((((..(((...((((((((.....))))...(((....)))))))-.)))...)))))).))).)))..----------- ( -34.26) >DroYak_CAF1 24070 115 - 1 AAGGGCUCCACAGCCCCAACCCUGAAAUGAAACACAGUCAUUGUGUCUUGGCCUGGCCAAAAACUGGCAUACCAAAAUGG----G-GGGAAAUCAGUGAAAGGCUUGACUCGGAAACAGG ..(((((....)))))....((((.(((((.......)))))(.(((..(((((.((((.....))))...((.....))----.-..............))))).))).).....)))) ( -31.90) >consensus AAGGGCUCCACAGCCCCAACCCUGAAAUGAAACACAGUCAUUGUGUCUUAGCCUGGCCAAAAACUGGCAUACCAAAAUGG____G_GGGAAAUCAGUGAAAGGCUUGACUCGGAAACAGG ..(((((....)))))(((.(((..............((((((..((((......((((.....))))...((.....))......))))...)))))).))).))).((((....)))) (-23.87 = -24.56 + 0.69)

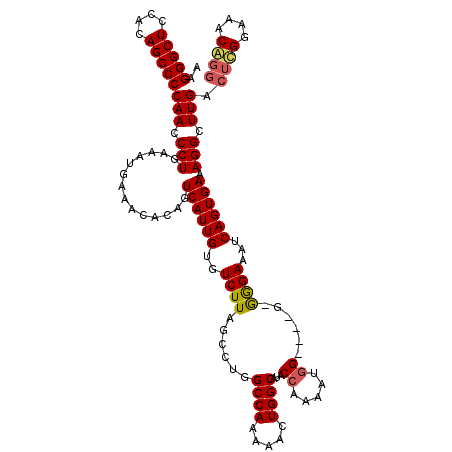
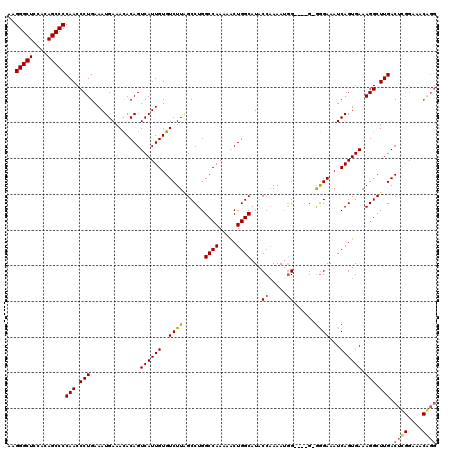
| Location | 18,779,417 – 18,779,535 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 95.62 |
| Mean single sequence MFE | -36.83 |
| Consensus MFE | -35.67 |
| Energy contribution | -35.42 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751182 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18779417 118 + 22224390 CCUUUUUGGCAUGCCAGUUUUUGGCCAGGCUAAGACACAAUGACUGUAUUUCAUUUCAGGGUUGGGGCUGCGGAGCCCUUUUCUUCUUCGAGGAACCCUCGCAGAAAUUGUCAAACAA ...(((((((..(((((...)))))...))))))).....((((...(((((.....((((..((((((....))))))..))))...(((((...)))))..))))).))))..... ( -37.00) >DroSec_CAF1 25910 118 + 1 CUAUUUUGGCAUGCCAGCUUUUGGCCAGGCUAAGACACAAUGACUGUGUUUCAUUUCAGGGUUGGGGCUGCGGAGCCCUUUUUUUCUUCGAGGAACCCUCGCAGAAAUUGUCAAACAA .....(((((..(((((...)))))...)))))((((....((((.((........)).))))((((((....))))))...(((((.(((((...))))).))))).))))...... ( -37.20) >DroEre_CAF1 24411 118 + 1 CCAUUUGGGUAUGCCAGUUUUUGGCCAGGCAAAGACACAAUGACUGUGUUUCAUUUCAGGGUUGGGGCUGUGGAGCCCUUUUCUUCUUCGAGGAACCCUCGCAGAAAUUGUCAAACAA ((....))....(((((...)))))..((((((((((((.....))))))).......(((..((((((....))))))..)))(((.(((((...))))).)))..)))))...... ( -35.00) >DroYak_CAF1 24105 118 + 1 CCAUUUUGGUAUGCCAGUUUUUGGCCAGGCCAAGACACAAUGACUGUGUUUCAUUUCAGGGUUGGGGCUGUGGAGCCCUUUUCUUCUUCGAGGAACCCUCGCAGAAAUUGUCAAACAA ...(((((((..(((((...)))))...))))))).....((((...(((((.....((((..((((((....))))))..))))...(((((...)))))..))))).))))..... ( -38.10) >consensus CCAUUUUGGCAUGCCAGUUUUUGGCCAGGCUAAGACACAAUGACUGUGUUUCAUUUCAGGGUUGGGGCUGCGGAGCCCUUUUCUUCUUCGAGGAACCCUCGCAGAAAUUGUCAAACAA ...(((((((..(((((...)))))...))))))).....((((...(((((.....((((..((((((....))))))..))))...(((((...)))))..))))).))))..... (-35.67 = -35.42 + -0.25)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:54 2006