| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,775,787 – 18,775,986 |
| Length | 199 |
| Max. P | 0.981696 |

| Location | 18,775,787 – 18,775,907 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.50 |
| Mean single sequence MFE | -29.43 |
| Consensus MFE | -19.64 |
| Energy contribution | -20.20 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.618380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18775787 120 + 22224390 CCUGAGCAAGGCGAACUCAAGUCCAAACUUCUACUCCUGAGCGACUUUCCUCUUUUCCGGAGACCAUUCAGAGGAUAAGGUCAGGAAAACGCUUUACAAAUCCUUGAAAGAGACUCGGAA .(((((.((((((.....((((....))))....((((((.(.....((((((.....(....).....))))))...).))))))...)))))).......(((....))).))))).. ( -34.70) >DroSec_CAF1 21093 120 + 1 CCUGAGGAAGGUGAACUGUAGUCCAAACUCCUACUCCUCAGCGACUUUCCUCUUUUCCGGAGACCAUUCAGAGGAUAAGGUCAGGAAAACGCUUUACAAAUCCUCGAAAGAGACUCGGAA .(((((.((((((....((((.........))))((((....(((((((((((.....(....).....))))))..)))))))))...)))))).......(((....))).))))).. ( -35.10) >DroEre_CAF1 21034 107 + 1 CCUUAGGAAUUCCAAC-------------UAUAGUCCUAAGCGACUUUCCUCUUUUCCGGAGUCCAUUCGGAGGAUAAGGUCAGGAAAACGCUUUACAAAUCCUCGAAACAGACGCGGAA .(((((((........-------------.....))))))).............(((((..(((..((((.(((((...((.(((......))).))..)))))))))...))).))))) ( -26.22) >DroYak_CAF1 20810 93 + 1 CU---------------------------CAUCCUCCUAAGCGACUUUUCUCUUUUCCGGAGUUCAUUCAGAGGAUAAGGUCAGGAAAACGCUUUACAAAUCCUCGAAAGAGACUCGGAA ..---------------------------..(((....(((((..(((((((((.(((.(((....)))...))).)))...)))))).)))))........(((....)))....))). ( -21.70) >consensus CCUGAGGAAGGCGAAC_____________CAUACUCCUAAGCGACUUUCCUCUUUUCCGGAGACCAUUCAGAGGAUAAGGUCAGGAAAACGCUUUACAAAUCCUCGAAAGAGACUCGGAA ..................................(((.(((((..(((((((((.(((.(((....)))...))).)))...)))))).)))))........(((....)))....))). (-19.64 = -20.20 + 0.56)



| Location | 18,775,827 – 18,775,947 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.70 |
| Mean single sequence MFE | -35.30 |
| Consensus MFE | -32.41 |
| Energy contribution | -33.10 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18775827 120 + 22224390 GCGACUUUCCUCUUUUCCGGAGACCAUUCAGAGGAUAAGGUCAGGAAAACGCUUUACAAAUCCUUGAAAGAGACUCGGAAAAGCGGAUGGCGAAAAAAAUCGCCGAAAAUGAAACGAAGC .((....(((.((((((((.((.((.((((.(((((...((.(((......))).))..))))))))).).).)))))))))).)))((((((......)))))).........)).... ( -34.90) >DroSec_CAF1 21133 119 + 1 GCGACUUUCCUCUUUUCCGGAGACCAUUCAGAGGAUAAGGUCAGGAAAACGCUUUACAAAUCCUCGAAAGAGACUCGGAAAAGCGGAUGGCGAAA-AAAUCGCCGAAAAUGAAGCGAAGC ((.....(((.((((((((.((.((.(((.((((((...((.(((......))).))..))))))))).).).)))))))))).)))((((((..-...))))))........))..... ( -38.20) >DroEre_CAF1 21061 118 + 1 GCGACUUUCCUCUUUUCCGGAGUCCAUUCGGAGGAUAAGGUCAGGAAAACGCUUUACAAAUCCUCGAAACAGACGCGGAAAAGCGGAUGGGGAA--AAAUCGCCGAAAAUGAAACGAAGC .((....(((.((((((((..(((..((((.(((((...((.(((......))).))..)))))))))...))).)))))))).)))(((.((.--...)).))).........)).... ( -34.90) >DroYak_CAF1 20823 119 + 1 GCGACUUUUCUCUUUUCCGGAGUUCAUUCAGAGGAUAAGGUCAGGAAAACGCUUUACAAAUCCUCGAAAGAGACUCGGAAAAGCGGAUGGGGAAA-AAAUCGCCGAAAAUGAAACCAAGC ((.....(((.(((((((((..(((.(((.((((((...((.(((......))).))..))))))))).)))..))))))))).)))(((.((..-...)).))).............)) ( -33.20) >consensus GCGACUUUCCUCUUUUCCGGAGACCAUUCAGAGGAUAAGGUCAGGAAAACGCUUUACAAAUCCUCGAAAGAGACUCGGAAAAGCGGAUGGCGAAA_AAAUCGCCGAAAAUGAAACGAAGC ((.....(((.((((((((.((((..((..((((((...((.(((......))).))..))))))..))..)))))))))))).)))((((((......)))))).............)) (-32.41 = -33.10 + 0.69)


| Location | 18,775,827 – 18,775,947 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.70 |
| Mean single sequence MFE | -37.03 |
| Consensus MFE | -31.77 |
| Energy contribution | -33.77 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18775827 120 - 22224390 GCUUCGUUUCAUUUUCGGCGAUUUUUUUCGCCAUCCGCUUUUCCGAGUCUCUUUCAAGGAUUUGUAAAGCGUUUUCCUGACCUUAUCCUCUGAAUGGUCUCCGGAAAAGAGGAAAGUCGC ((((............(((((......))))).(((.((((((((((.((..(((((((((.....(((.(((.....))))))))))).)))).)).)).)))))))).)))))))... ( -37.20) >DroSec_CAF1 21133 119 - 1 GCUUCGCUUCAUUUUCGGCGAUUU-UUUCGCCAUCCGCUUUUCCGAGUCUCUUUCGAGGAUUUGUAAAGCGUUUUCCUGACCUUAUCCUCUGAAUGGUCUCCGGAAAAGAGGAAAGUCGC ....(((((.......(((((...-..))))).(((.((((((((((.((.(((.((((((.....(((.(((.....)))))))))))).))).)).)).)))))))).)))))).)). ( -39.50) >DroEre_CAF1 21061 118 - 1 GCUUCGUUUCAUUUUCGGCGAUUU--UUCCCCAUCCGCUUUUCCGCGUCUGUUUCGAGGAUUUGUAAAGCGUUUUCCUGACCUUAUCCUCCGAAUGGACUCCGGAAAAGAGGAAAGUCGC .................(((((((--.......(((.((((((((.((((((((.((((((.....(((.(((.....)))))))))))).))))))))..)))))))).)))))))))) ( -38.71) >DroYak_CAF1 20823 119 - 1 GCUUGGUUUCAUUUUCGGCGAUUU-UUUCCCCAUCCGCUUUUCCGAGUCUCUUUCGAGGAUUUGUAAAGCGUUUUCCUGACCUUAUCCUCUGAAUGAACUCCGGAAAAGAGAAAAGUCGC .................(((((((-(((.........(((((((((((.((.(((((((((.....(((.(((.....)))))))))))).))).))))).)))))))).)))))))))) ( -32.70) >consensus GCUUCGUUUCAUUUUCGGCGAUUU_UUUCCCCAUCCGCUUUUCCGAGUCUCUUUCGAGGAUUUGUAAAGCGUUUUCCUGACCUUAUCCUCUGAAUGGACUCCGGAAAAGAGGAAAGUCGC ((((............(((((......))))).(((.((((((((((((..(((.((((((.....(((.(((.....)))))))))))).)))..)))).)))))))).)))))))... (-31.77 = -33.77 + 2.00)


| Location | 18,775,867 – 18,775,986 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 95.36 |
| Mean single sequence MFE | -26.61 |
| Consensus MFE | -20.83 |
| Energy contribution | -21.95 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18775867 119 + 22224390 UCAGGAAAACGCUUUACAAAUCCUUGAAAGAGACUCGGAAAAGCGGAUGGCGAAAAAAAUCGCCGAAAAUGAAACGAAGCGAAAAUAAAAUUUCCCAGCUAAUGACAAAAAUUUCCGGU ...(((((.((((((.(.....(((....))).....).))))))..((((((......))))))............(((((((......))))...)))...........)))))... ( -24.90) >DroSec_CAF1 21173 118 + 1 UCAGGAAAACGCUUUACAAAUCCUCGAAAGAGACUCGGAAAAGCGGAUGGCGAAA-AAAUCGCCGAAAAUGAAGCGAAGCGAAAAUAAAAUUUCCAAGCUAAUGACAAAAAUUUCCGGU ...(((((.((((((.(.....(((....))).....).))))))..((((((..-...)))))).......(((...(.((((......)))))..)))...........)))))... ( -29.70) >DroEre_CAF1 21101 117 + 1 UCAGGAAAACGCUUUACAAAUCCUCGAAACAGACGCGGAAAAGCGGAUGGGGAA--AAAUCGCCGAAAAUGAAACGAAGCGAACAUAAAAUUUCCCAGCUAAUGACAAAAAUUUCCGGU ...(((((.((((((.....(((.((.......)).)))))))))..(((((((--...((((((.........))..))))........)))))))..............)))))... ( -24.20) >DroYak_CAF1 20863 118 + 1 UCAGGAAAACGCUUUACAAAUCCUCGAAAGAGACUCGGAAAAGCGGAUGGGGAAA-AAAUCGCCGAAAAUGAAACCAAGCGAAUAUAAAAUUUCCCAGCUAAUGACAAAAAUUUCCGGU ...(((((.((((((.(.....(((....))).....).))))))..(((((((.-...((((...............))))........)))))))..............)))))... ( -27.66) >consensus UCAGGAAAACGCUUUACAAAUCCUCGAAAGAGACUCGGAAAAGCGGAUGGCGAAA_AAAUCGCCGAAAAUGAAACGAAGCGAAAAUAAAAUUUCCCAGCUAAUGACAAAAAUUUCCGGU ...(((((.((((((.(.....(((....))).....).))))))..((((((......))))))............(((.................)))...........)))))... (-20.83 = -21.95 + 1.13)


| Location | 18,775,867 – 18,775,986 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 95.36 |
| Mean single sequence MFE | -27.05 |
| Consensus MFE | -22.54 |
| Energy contribution | -23.10 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18775867 119 - 22224390 ACCGGAAAUUUUUGUCAUUAGCUGGGAAAUUUUAUUUUCGCUUCGUUUCAUUUUCGGCGAUUUUUUUCGCCAUCCGCUUUUCCGAGUCUCUUUCAAGGAUUUGUAAAGCGUUUUCCUGA ...(((((....((.....(((.(..((......))..))))......)).....(((((......)))))...((((((..(((((((.......))))))).)))))).)))))... ( -28.50) >DroSec_CAF1 21173 118 - 1 ACCGGAAAUUUUUGUCAUUAGCUUGGAAAUUUUAUUUUCGCUUCGCUUCAUUUUCGGCGAUUU-UUUCGCCAUCCGCUUUUCCGAGUCUCUUUCGAGGAUUUGUAAAGCGUUUUCCUGA ...(((((...........(((..(((((......)))).)...)))........(((((...-..)))))...((((((..((((((((....)).)))))).)))))).)))))... ( -28.10) >DroEre_CAF1 21101 117 - 1 ACCGGAAAUUUUUGUCAUUAGCUGGGAAAUUUUAUGUUCGCUUCGUUUCAUUUUCGGCGAUUU--UUCCCCAUCCGCUUUUCCGCGUCUGUUUCGAGGAUUUGUAAAGCGUUUUCCUGA ...(((((...............((((((........(((((.............)))))..)--)))))....(((((((((.((.......)).))).....)))))).)))))... ( -23.82) >DroYak_CAF1 20863 118 - 1 ACCGGAAAUUUUUGUCAUUAGCUGGGAAAUUUUAUAUUCGCUUGGUUUCAUUUUCGGCGAUUU-UUUCCCCAUCCGCUUUUCCGAGUCUCUUUCGAGGAUUUGUAAAGCGUUUUCCUGA ...(((((...............((((((........(((((.((........)))))))...-))))))....((((((..((((((((....)).)))))).)))))).)))))... ( -27.80) >consensus ACCGGAAAUUUUUGUCAUUAGCUGGGAAAUUUUAUUUUCGCUUCGUUUCAUUUUCGGCGAUUU_UUUCCCCAUCCGCUUUUCCGAGUCUCUUUCGAGGAUUUGUAAAGCGUUUUCCUGA ...(((((...........(((..((..(........)..))..)))........(((((......)))))...((((((..(((((((.......))))))).)))))).)))))... (-22.54 = -23.10 + 0.56)

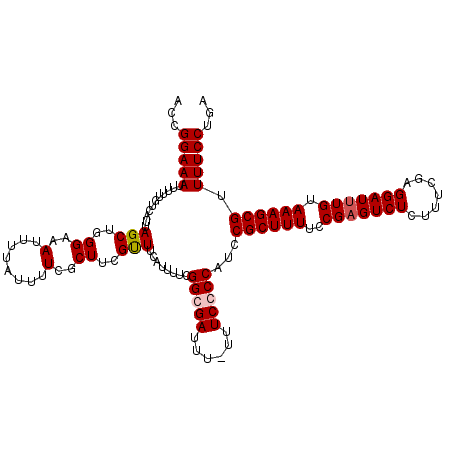
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:49 2006