| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,740,847 – 18,740,944 |
| Length | 97 |
| Max. P | 0.994627 |

| Location | 18,740,847 – 18,740,944 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 72.76 |
| Mean single sequence MFE | -21.74 |
| Consensus MFE | -7.42 |
| Energy contribution | -8.28 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.614663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
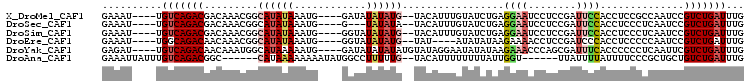
>X_DroMel_CAF1 18740847 97 + 22224390 GAAAU----UGUCAGACGACAAACGGCAUAUAAAUG----GAUAUAUAUG--UACAUUUGUAUCUGAGGAAUCCUCCGAUUCCACCUCCGCCAAUCCGUCUGAUUUG .....----.((((((((......((((((((((((----.(((....))--).)))))))))....((((((....))))))......)))....))))))))... ( -28.40) >DroSec_CAF1 808 94 + 1 GAAAU----UGUCAGACGACAAACGGCAUAUAAAUG----G---UAUAUA--UACAUUUGUAUCUGAGGAAUCCUCCGAUUCCACCUCCCUCAAUCCGUCUGAUUUG .....----.((((((((.....(((.(((((((((----.---......--..)))))))))))).((((((....)))))).............))))))))... ( -28.40) >DroSim_CAF1 8419 97 + 1 GAAAU----UGUCAGACGACAAACGGCAUAUAAAUG----GGUAUAUAUG--UACAUUUGUAUCUGAGGAAUCCUCCGAUUCCACCUCCCUCAAUCCGUCUGAUUUG .....----.((((((((.....(((.(((((((((----..((....))--..)))))))))))).((((((....)))))).............))))))))... ( -28.60) >DroEre_CAF1 8860 93 + 1 GAAAU----UGGCAGACAACAAACGGCAUAUAAAUG----GGUAUAUAUG--UAU----AUAUAUAAGAAAACCUCCGAUCCCACCUCCCCCAAUCCGUCUGAUUUG .....----...(((((..(....).........((----(((((((((.--...----))))))).((.....)).............))))....)))))..... ( -11.40) >DroYak_CAF1 9802 99 + 1 GAGAU----UGUCAGACAACAAAUGGCAUAAAAAUG----GAUAUAUAUAUGUAUAGGAAUAUAUAAGAAACCCAGCGAUUUCACCCCCCUCAAUUCGUCUGAUUUG .....----.(((((((...................----....(((((((........))))))).((((........))))..............)))))))... ( -14.00) >DroAna_CAF1 8147 93 + 1 GAAAUUAUUUGUCAGACGGC------CAUAAAAAAAAUAUGGCCUUUUUG--UACAUUUUUUUUAUUGGU------UUAUUUUAUUUUCCCGCUGCUGUCUGAUUUG ..........((((((((((------.(((((((((((...((......)--)..))))))))))).((.------.............))...))))))))))... ( -19.64) >consensus GAAAU____UGUCAGACGACAAACGGCAUAUAAAUG____GGUAUAUAUG__UACAUUUGUAUCUGAGGAAUCCUCCGAUUCCACCUCCCCCAAUCCGUCUGAUUUG ..........(((((((.........((((((............)))))).................((((........))))..............)))))))... ( -7.42 = -8.28 + 0.86)
| Location | 18,740,847 – 18,740,944 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 72.76 |
| Mean single sequence MFE | -25.61 |
| Consensus MFE | -13.76 |
| Energy contribution | -14.73 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.54 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994627 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
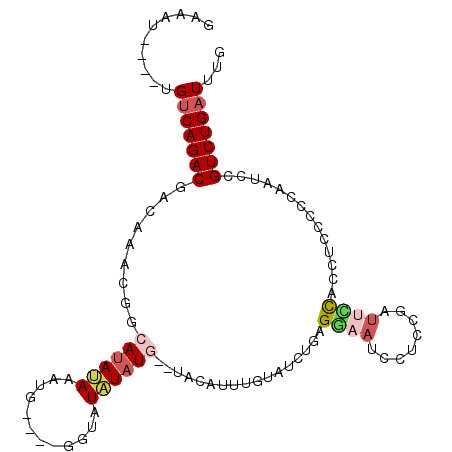
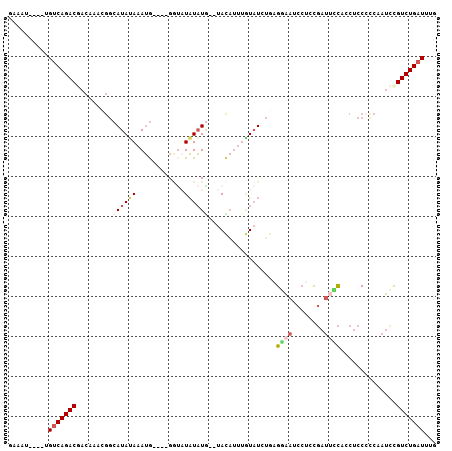
>X_DroMel_CAF1 18740847 97 - 22224390 CAAAUCAGACGGAUUGGCGGAGGUGGAAUCGGAGGAUUCCUCAGAUACAAAUGUA--CAUAUAUAUC----CAUUUAUAUGCCGUUUGUCGUCUGACA----AUUUC ....((((((((..(((((.((((((((((.((((...)))).)))....((((.--...)))).))----)))))...)))))....))))))))..----..... ( -31.30) >DroSec_CAF1 808 94 - 1 CAAAUCAGACGGAUUGAGGGAGGUGGAAUCGGAGGAUUCCUCAGAUACAAAUGUA--UAUAUA---C----CAUUUAUAUGCCGUUUGUCGUCUGACA----AUUUC ....(((((((...........(.((((((....)))))).)....(((((((((--((((..---.----....)))))).))))))))))))))..----..... ( -27.20) >DroSim_CAF1 8419 97 - 1 CAAAUCAGACGGAUUGAGGGAGGUGGAAUCGGAGGAUUCCUCAGAUACAAAUGUA--CAUAUAUACC----CAUUUAUAUGCCGUUUGUCGUCUGACA----AUUUC ....(((((((...........(.((((((....)))))).)....(((((((..--((((((....----....)))))).))))))))))))))..----..... ( -27.40) >DroEre_CAF1 8860 93 - 1 CAAAUCAGACGGAUUGGGGGAGGUGGGAUCGGAGGUUUUCUUAUAUAU----AUA--CAUAUAUACC----CAUUUAUAUGCCGUUUGUUGUCUGCCA----AUUUC .....(((((((..(((.(.(((((((...((((...))))(((((((----...--.)))))))))----)))))...).)))....)))))))...----..... ( -21.50) >DroYak_CAF1 9802 99 - 1 CAAAUCAGACGAAUUGAGGGGGGUGAAAUCGCUGGGUUUCUUAUAUAUUCCUAUACAUAUAUAUAUC----CAUUUUUAUGCCAUUUGUUGUCUGACA----AUCUC ....((((((((...((..((.((((((....(((((....(((((((........))))))).)))----)).)))))).)).))..))))))))..----..... ( -24.90) >DroAna_CAF1 8147 93 - 1 CAAAUCAGACAGCAGCGGGAAAAUAAAAUAA------ACCAAUAAAAAAAAUGUA--CAAAAAGGCCAUAUUUUUUUUAUG------GCCGUCUGACAAAUAAUUUC ....((((((......((.............------.))...............--......(((((((.......))))------)))))))))........... ( -21.34) >consensus CAAAUCAGACGGAUUGAGGGAGGUGGAAUCGGAGGAUUCCUCAGAUACAAAUGUA__CAUAUAUACC____CAUUUAUAUGCCGUUUGUCGUCUGACA____AUUUC ....((((((((...((.((....((((((....)))))).................((((((............)))))))).))..))))))))........... (-13.76 = -14.73 + 0.98)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:43 2006