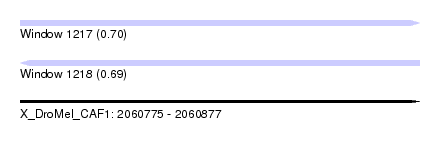
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,060,775 – 2,060,877 |
| Length | 102 |
| Max. P | 0.704379 |

| Location | 2,060,775 – 2,060,877 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 80.44 |
| Mean single sequence MFE | -38.02 |
| Consensus MFE | -29.04 |
| Energy contribution | -29.60 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2060775 102 + 22224390 ACCUGUUGGGACUGGACAAGCUGACCAACGCCUUUGGCCUGCUGCUGCUCUUCCAGGGUAUUGCCAGCUUCAUCGGUCCGCCCAUCGGAGGUGAGUACUC---GC----- ..(((.((((...((((((((((..(((..((((.((...((....))....))))))..))).)))))).....)))).)))).)))..((((....))---))----- ( -34.40) >DroVir_CAF1 2987 105 + 1 AUCUGCUGGGCCUGGAGAAGCUGACCAAUGCCUUUGGUCUGCUGCUGCUCUUCCAGGGCAUUGCCAGUUUCAUUGGACCGCCCAUUGGCGGUGAGUUGCAUAUGC----- ...(((.....(..(.(((((((..(((((((((.((...((....))....))))))))))).))))))).)..)((((((....)))))).....))).....----- ( -40.60) >DroPse_CAF1 3187 107 + 1 AUUUGUUGGGGCUGGAGAAGCUGACCAAUGCCUUUGGCCUGCUGCUGCUCUUCCAGGGCAUUGCUAGCUUCAUUGGUCCCCCCAUCGGAGGUAAGUCUUU---CCCUCGU .......(((((..(.(((((((..(((((((((.((...((....))....))))))))))).))))))).)..))))).......((((.........---.)))).. ( -41.70) >DroGri_CAF1 2468 102 + 1 AUCUGUUGGGCCUGGACAAGCUGACCAAUGCAUUCGGGCUGCUCUUACUCUUCCAGGGCGUUGCCACGUUAAUUGGCCCGCCCAUAGGCGGUAAGAUUUG---GG----- ........((((((((...((........)).))))))))..((((((((((...(((((..((((.......)))).)))))..))).)))))))....---..----- ( -34.00) >DroMoj_CAF1 3150 102 + 1 AUCUGCUGGGCCUGGAGAAGCUGACCAAUGCGUUUGGACUGCUGCUGCUCUUCCAGGGCAUCGCUAGCUUCAUCGGACCACCCAUUGGAGGUGAGCUGCA---GU----- ..((((..((((((..(((((((.(..((((.((((((..((....))...)))))))))).).)))))))..)))..(((((....).)))).))))))---).----- ( -35.70) >DroPer_CAF1 3176 107 + 1 AUUUGUUGGGGCUGGAGAAGCUGACCAAUGCCUUUGGCCUGCUGCUGCUCUUCCAGGGCAUUGCUAGCUUCAUUGGUCCCCCCAUCGGAGGUAAGUCUUU---CCCUCGU .......(((((..(.(((((((..(((((((((.((...((....))....))))))))))).))))))).)..))))).......((((.........---.)))).. ( -41.70) >consensus AUCUGUUGGGCCUGGAGAAGCUGACCAAUGCCUUUGGCCUGCUGCUGCUCUUCCAGGGCAUUGCCAGCUUCAUUGGUCCGCCCAUCGGAGGUAAGUUUUA___GC_____ ..(((.(((((((((.(((((((..(((((((((.((...((....))....))))))))))).))))))).))))...))))).)))...................... (-29.04 = -29.60 + 0.56)

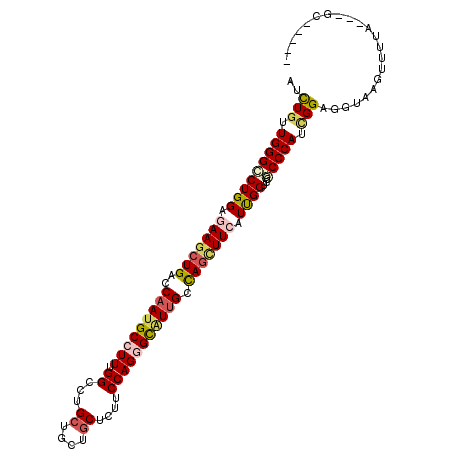
| Location | 2,060,775 – 2,060,877 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 80.44 |
| Mean single sequence MFE | -35.83 |
| Consensus MFE | -23.70 |
| Energy contribution | -24.07 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2060775 102 - 22224390 -----GC---GAGUACUCACCUCCGAUGGGCGGACCGAUGAAGCUGGCAAUACCCUGGAAGAGCAGCAGCAGGCCAAAGGCGUUGGUCAGCUUGUCCAGUCCCAACAGGU -----..---........((((..(.(((((((((.....((((((((...(((((((....((....))...))..))).))..)))))))))))).).)))).))))) ( -31.50) >DroVir_CAF1 2987 105 - 1 -----GCAUAUGCAACUCACCGCCAAUGGGCGGUCCAAUGAAACUGGCAAUGCCCUGGAAGAGCAGCAGCAGACCAAAGGCAUUGGUCAGCUUCUCCAGGCCCAGCAGAU -----((((.(((.....((((((....)))))).((.......)))))))))((((((..(((.(...).((((((.....)))))).)))..)))))).......... ( -36.30) >DroPse_CAF1 3187 107 - 1 ACGAGGG---AAAGACUUACCUCCGAUGGGGGGACCAAUGAAGCUAGCAAUGCCCUGGAAGAGCAGCAGCAGGCCAAAGGCAUUGGUCAGCUUCUCCAGCCCCAACAAAU ..((((.---.........)))).(.((((((((.....(((((((.(((((((.(((....((....))...)))..))))))).).)))))))))..))))).).... ( -38.40) >DroGri_CAF1 2468 102 - 1 -----CC---CAAAUCUUACCGCCUAUGGGCGGGCCAAUUAACGUGGCAACGCCCUGGAAGAGUAAGAGCAGCCCGAAUGCAUUGGUCAGCUUGUCCAGGCCCAACAGAU -----..---...((((....(((....)))(((((.......(((....)))..((((.((((..((.((((......))..)).)).)))).)))))))))...)))) ( -32.70) >DroMoj_CAF1 3150 102 - 1 -----AC---UGCAGCUCACCUCCAAUGGGUGGUCCGAUGAAGCUAGCGAUGCCCUGGAAGAGCAGCAGCAGUCCAAACGCAUUGGUCAGCUUCUCCAGGCCCAGCAGAU -----.(---(((.(((((.......)))))((((.(..(((((((.((((((..((((...((....))..))))...)))))).).))))))..).))))..)))).. ( -37.70) >DroPer_CAF1 3176 107 - 1 ACGAGGG---AAAGACUUACCUCCGAUGGGGGGACCAAUGAAGCUAGCAAUGCCCUGGAAGAGCAGCAGCAGGCCAAAGGCAUUGGUCAGCUUCUCCAGCCCCAACAAAU ..((((.---.........)))).(.((((((((.....(((((((.(((((((.(((....((....))...)))..))))))).).)))))))))..))))).).... ( -38.40) >consensus _____GC___AAAAACUCACCUCCGAUGGGCGGACCAAUGAAGCUAGCAAUGCCCUGGAAGAGCAGCAGCAGGCCAAAGGCAUUGGUCAGCUUCUCCAGGCCCAACAGAU ..........................((((((((.....(((((((.(((((((.(((....((....))...)))..))))))).).)))))))))..)))))...... (-23.70 = -24.07 + 0.37)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:55 2006