
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,730,957 – 18,731,066 |
| Length | 109 |
| Max. P | 0.972635 |

| Location | 18,730,957 – 18,731,066 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.64 |
| Mean single sequence MFE | -34.24 |
| Consensus MFE | -27.15 |
| Energy contribution | -28.19 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18730957 109 + 22224390 -----------AUUUAAAGCAAACAAAUUUAUUGCCGUGCAGGCCAGGAGAUUUGUUUUCCAAUUGGCCUGGAGCAGGAGCAGGAGCAGGAGCAUUGGAGGGCAGUGAAUGAAGCCAACU -----------.......((......((((((((((((.((((((((((((.....)))))...)))))))..)).....(((..((....)).)))...))))))))))...))..... ( -33.40) >DroSec_CAF1 3860 103 + 1 -----------GUUUAAGGCAAACAAAUUUAUUGCCGUGCAGGCCAGGAGAUUUGUUUUCCAAUUGGCCUGGAGCAGGA------GCAGGAGCAUUGGAGGGCAGUGAAUGAAGCCAACU -----------......(((......((((((((((((.((((((((((((.....)))))...)))))))..))....------((....)).......))))))))))...))).... ( -37.10) >DroSim_CAF1 3921 103 + 1 -----------AUUUAAGGCAAACAAAUUUAUUGCCGUGCAGGCCAGGAGAUUUGUUUUCCAAUUGGCCUGGAGCAGGA------GCAGGAGCAUUGGAGGGCAGUGAAUGAAGCCAACU -----------......(((......((((((((((((.((((((((((((.....)))))...)))))))..))....------((....)).......))))))))))...))).... ( -37.10) >DroEre_CAF1 1866 108 + 1 UCUAUAGAUCUUAUUGAAGCACUCAAAUUUCUUGCCGUGCAGGCAAGGGGAUUUGUUUUCCAAUUGGCCUGGAGAA------------GGAGCAUUGGAGGGCAGUGAAUGAAGCCAACU .............(((...((.(((..(..((((((.....))))))..)..((((((((((((...(((.....)------------))...))))))))))))))).))....))).. ( -31.50) >DroYak_CAF1 3219 108 + 1 UUGAUAGUUAUUUUUGAGGCACUCAAAUUUAUUGCCGUGCAGGCAAGGAGAUUUGUUUUCCAAUUGGCCUGGAGCA------------GGAGCAUUGGAGGGCAGUGAAUGAAGCCAACU .................(((..(((..(((((((((.(.(((((..(((((.....)))))......((((...))------------)).)).))).).)))))))))))).))).... ( -32.10) >consensus ___________AUUUAAGGCAAACAAAUUUAUUGCCGUGCAGGCCAGGAGAUUUGUUUUCCAAUUGGCCUGGAGCAGGA______GCAGGAGCAUUGGAGGGCAGUGAAUGAAGCCAACU .................(((......((((((((((...((((((((((((.....)))))...)))))))..((................)).......))))))))))...))).... (-27.15 = -28.19 + 1.04)

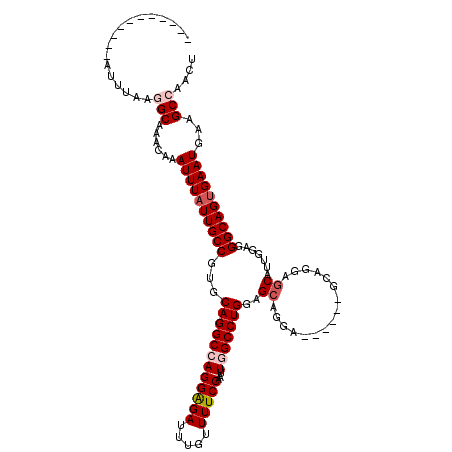

| Location | 18,730,957 – 18,731,066 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.64 |
| Mean single sequence MFE | -24.55 |
| Consensus MFE | -17.41 |
| Energy contribution | -17.61 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716717 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18730957 109 - 22224390 AGUUGGCUUCAUUCACUGCCCUCCAAUGCUCCUGCUCCUGCUCCUGCUCCAGGCCAAUUGGAAAACAAAUCUCCUGGCCUGCACGGCAAUAAAUUUGUUUGCUUUAAAU----------- ....(((..........))).((((((...((((.....((....))..))))...))))))((((((((......(((.....))).....)))))))).........----------- ( -25.20) >DroSec_CAF1 3860 103 - 1 AGUUGGCUUCAUUCACUGCCCUCCAAUGCUCCUGC------UCCUGCUCCAGGCCAAUUGGAAAACAAAUCUCCUGGCCUGCACGGCAAUAAAUUUGUUUGCCUUAAAC----------- ....(((..............((((((...((((.------........))))...))))))((((((((......(((.....))).....)))))))))))......----------- ( -24.20) >DroSim_CAF1 3921 103 - 1 AGUUGGCUUCAUUCACUGCCCUCCAAUGCUCCUGC------UCCUGCUCCAGGCCAAUUGGAAAACAAAUCUCCUGGCCUGCACGGCAAUAAAUUUGUUUGCCUUAAAU----------- ....(((..............((((((...((((.------........))))...))))))((((((((......(((.....))).....)))))))))))......----------- ( -24.20) >DroEre_CAF1 1866 108 - 1 AGUUGGCUUCAUUCACUGCCCUCCAAUGCUCC------------UUCUCCAGGCCAAUUGGAAAACAAAUCCCCUUGCCUGCACGGCAAGAAAUUUGAGUGCUUCAAUAAGAUCUAUAGA .(((((...((((((......((((((...((------------(.....)))...))))))...........((((((.....)))))).....))))))..)))))............ ( -25.10) >DroYak_CAF1 3219 108 - 1 AGUUGGCUUCAUUCACUGCCCUCCAAUGCUCC------------UGCUCCAGGCCAAUUGGAAAACAAAUCUCCUUGCCUGCACGGCAAUAAAUUUGAGUGCCUCAAAAAUAACUAUCAA (((((........((((....((((((...((------------((...))))...))))))...(((((....(((((.....)))))...))))))))).........)))))..... ( -24.03) >consensus AGUUGGCUUCAUUCACUGCCCUCCAAUGCUCCUGC______UCCUGCUCCAGGCCAAUUGGAAAACAAAUCUCCUGGCCUGCACGGCAAUAAAUUUGUUUGCCUUAAAA___________ ....(((..((.....((((.......((................))..(((((((...(((.......)))..)))))))...)))).......))...)))................. (-17.41 = -17.61 + 0.20)

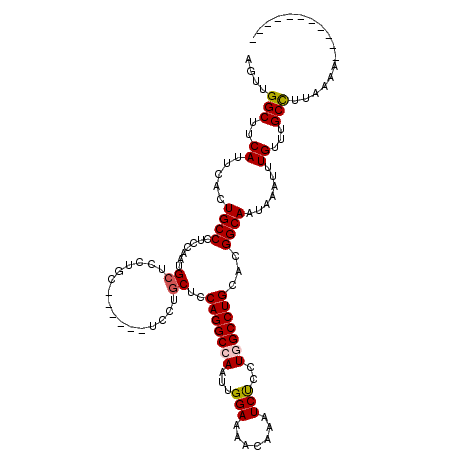
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:41 2006