
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,715,070 – 18,715,221 |
| Length | 151 |
| Max. P | 0.998761 |

| Location | 18,715,070 – 18,715,181 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 77.71 |
| Mean single sequence MFE | -26.37 |
| Consensus MFE | -13.57 |
| Energy contribution | -14.13 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.26 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.961043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18715070 111 + 22224390 AGAAACCGAUUAUAAAGCGAAUAUAUAAAUGAAAUUACAUCGAAUCCAGACCGAUUUCGAUUUCGAUUUCGAUUUGGAUUUGGAUUUCGGACUCAGAUUCGAAUUCUGAAA .....(((((((((.........)))))...........(((((((((((.(((..(((....)))..))).)))))))))))...))))..(((((.......))))).. ( -30.00) >DroSec_CAF1 16388 92 + 1 AGAAACCGAUUAUAAAGCGAAUAUAUAAAUGAAAUUACAUCGAAUCCAGGCCGAUU------------------UGGAUUUGGAU-UCGGAUUCGGAUUCGAGUUCUGAAA .(((.(((((((((.........)))))...........(((((((((((((....------------------.)).)))))))-))))..)))).)))........... ( -22.30) >DroYak_CAF1 15970 90 + 1 AGAAUCCGAUUAUAAUGCG--UAUAUAAAUGAAAUUACAUCGAGUCCAUACCGAUU------------------UCGAUGUGGAU-UCAAAUUCGGAUUCGAGUUUUGUAA .((((((((.........(--((............)))...(((((((((.((...------------------.)).)))))))-))....))))))))........... ( -26.80) >consensus AGAAACCGAUUAUAAAGCGAAUAUAUAAAUGAAAUUACAUCGAAUCCAGACCGAUU__________________UGGAUUUGGAU_UCGGAUUCGGAUUCGAGUUCUGAAA .(((.(((((((((.........)))))...........((((((((((((((...((((........))))..))).))))))).))))..)))).)))........... (-13.57 = -14.13 + 0.56)

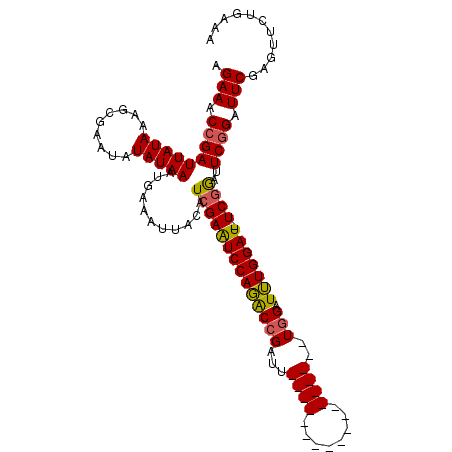

| Location | 18,715,070 – 18,715,181 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 77.71 |
| Mean single sequence MFE | -24.33 |
| Consensus MFE | -14.37 |
| Energy contribution | -14.59 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.55 |
| Structure conservation index | 0.59 |
| SVM decision value | 3.22 |
| SVM RNA-class probability | 0.998761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18715070 111 - 22224390 UUUCAGAAUUCGAAUCUGAGUCCGAAAUCCAAAUCCAAAUCGAAAUCGAAAUCGAAAUCGGUCUGGAUUCGAUGUAAUUUCAUUUAUAUAUUCGCUUUAUAAUCGGUUUCU .((((((.......))))))...(((((((.((((((.(((((..(((....)))..))))).)))))).)((((((......))))))...............)))))). ( -26.00) >DroSec_CAF1 16388 92 - 1 UUUCAGAACUCGAAUCCGAAUCCGA-AUCCAAAUCCA------------------AAUCGGCCUGGAUUCGAUGUAAUUUCAUUUAUAUAUUCGCUUUAUAAUCGGUUUCU ...........(((.((((...(((-(((((...((.------------------....))..))))))))((((((......)))))).............)))).))). ( -22.10) >DroYak_CAF1 15970 90 - 1 UUACAAAACUCGAAUCCGAAUUUGA-AUCCACAUCGA------------------AAUCGGUAUGGACUCGAUGUAAUUUCAUUUAUAUA--CGCAUUAUAAUCGGAUUCU ...........((((((((...(((-.((((.((((.------------------...)))).)))).)))((((((......)))))).--..........)))))))). ( -24.90) >consensus UUUCAGAACUCGAAUCCGAAUCCGA_AUCCAAAUCCA__________________AAUCGGUCUGGAUUCGAUGUAAUUUCAUUUAUAUAUUCGCUUUAUAAUCGGUUUCU ...........(((.((((...(((.(((((.(((........................))).))))))))((((((......)))))).............)))).))). (-14.37 = -14.59 + 0.23)

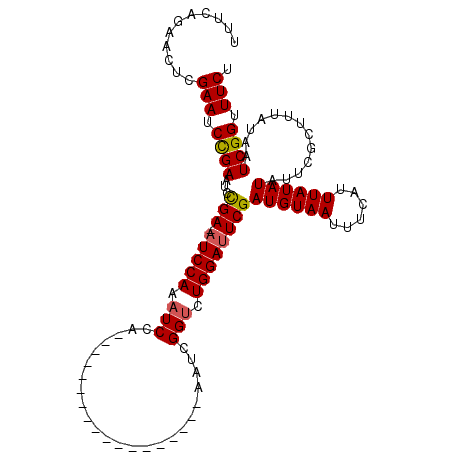
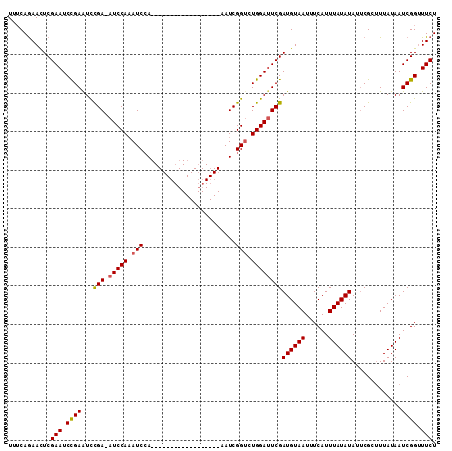
| Location | 18,715,101 – 18,715,221 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.23 |
| Mean single sequence MFE | -28.10 |
| Consensus MFE | -17.15 |
| Energy contribution | -17.60 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.608093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18715101 120 + 22224390 AAAUUACAUCGAAUCCAGACCGAUUUCGAUUUCGAUUUCGAUUUGGAUUUGGAUUUCGGACUCAGAUUCGAAUUCUGAAAGCAAUCGCGUAAUUUGUAAACUUUUAGUAGGCAGUGUUUU (((((((.(((((((((((.(((..(((....)))..))).))))))))))).(((((((.((......))..)))))))((....)))))))))((..((.....))..))........ ( -32.90) >DroSec_CAF1 16419 101 + 1 AAAUUACAUCGAAUCCAGGCCGAUU------------------UGGAUUUGGAU-UCGGAUUCGGAUUCGAGUUCUGAAAGCAAUCGCGUAAUUUGUAAACUUUUAGUAGGCAGUGUUUU (((((((.(((((((((((((....------------------.)).)))))))-)))).((((((.......)))))).((....)))))))))((..((.....))..))........ ( -26.80) >DroYak_CAF1 15999 101 + 1 AAAUUACAUCGAGUCCAUACCGAUU------------------UCGAUGUGGAU-UCAAAUUCGGAUUCGAGUUUUGUAAGCAAUCGCGUUAUUUGUAAACUUUUAGUAGGCAGUGUUUU ...(((((..(((((((((.((...------------------.)).)))))))-))(((((((....))))))))))))((....)).....((((..((.....))..))))...... ( -24.60) >consensus AAAUUACAUCGAAUCCAGACCGAUU__________________UGGAUUUGGAU_UCGGAUUCGGAUUCGAGUUCUGAAAGCAAUCGCGUAAUUUGUAAACUUUUAGUAGGCAGUGUUUU (((((((...((((((((((((...((((........))))..))).))))))).))(((((((....))))))).....((....)))))))))((..((.....))..))........ (-17.15 = -17.60 + 0.45)



| Location | 18,715,101 – 18,715,221 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.23 |
| Mean single sequence MFE | -21.12 |
| Consensus MFE | -15.18 |
| Energy contribution | -15.18 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18715101 120 - 22224390 AAAACACUGCCUACUAAAAGUUUACAAAUUACGCGAUUGCUUUCAGAAUUCGAAUCUGAGUCCGAAAUCCAAAUCCAAAUCGAAAUCGAAAUCGAAAUCGGUCUGGAUUCGAUGUAAUUU .........................((((((((((((((..((((((.......))))))..))).......(((((.(((((..(((....)))..))))).)))))))).)))))))) ( -27.50) >DroSec_CAF1 16419 101 - 1 AAAACACUGCCUACUAAAAGUUUACAAAUUACGCGAUUGCUUUCAGAACUCGAAUCCGAAUCCGA-AUCCAAAUCCA------------------AAUCGGCCUGGAUUCGAUGUAAUUU .........................(((((((((....)).........(((((((((...((((-...........------------------..))))..))))))))).))))))) ( -19.72) >DroYak_CAF1 15999 101 - 1 AAAACACUGCCUACUAAAAGUUUACAAAUAACGCGAUUGCUUACAAAACUCGAAUCCGAAUUUGA-AUCCACAUCGA------------------AAUCGGUAUGGACUCGAUGUAAUUU .....................(((((.....((.((((..............))))))...((((-.((((.((((.------------------...)))).)))).)))))))))... ( -16.14) >consensus AAAACACUGCCUACUAAAAGUUUACAAAUUACGCGAUUGCUUUCAGAACUCGAAUCCGAAUCCGA_AUCCAAAUCCA__________________AAUCGGUCUGGAUUCGAUGUAAUUU .........................(((((((((....)).........(((((((((...((((................................))))..))))))))).))))))) (-15.18 = -15.18 + 0.01)

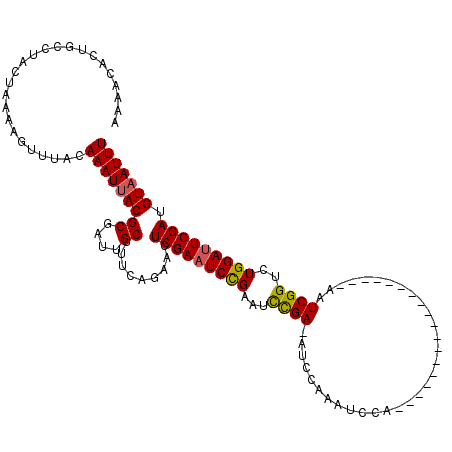

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:38 2006