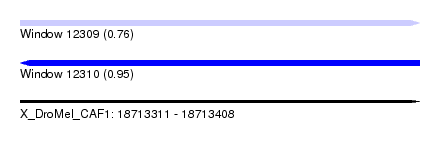
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,713,311 – 18,713,408 |
| Length | 97 |
| Max. P | 0.945703 |

| Location | 18,713,311 – 18,713,408 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 86.25 |
| Mean single sequence MFE | -34.84 |
| Consensus MFE | -20.09 |
| Energy contribution | -21.73 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.81 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
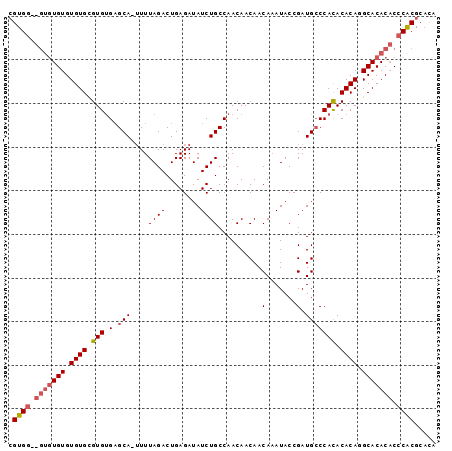
>X_DroMel_CAF1 18713311 97 + 22224390 CGUGGGUGUGUGUGUGUGCGUGUGAGCA-UUUUAGACUGAGAUAUCUGCCAACAACAACAAAUACCGAUGCCCACACACAGGCACACACCCACGCAUA (((((((((((((.((((.(((.(.(((-((.((((........))))..................))))))))).)))).))))))))))))).... ( -39.91) >DroSec_CAF1 14709 92 + 1 AGUGG-----UGUGUGUGCGUGUGAGCA-UUUUAGACUGAGAUAUCUGCCAACAACAACAAAUACCGAUGCCCACACACAGGCACACACCCACGCUCA .((((-----.(((((((((((((.(((-((.((((........))))..................))))).)))))....))))))))))))..... ( -30.31) >DroEre_CAF1 13918 94 + 1 CGUG---GUGUGUGUGUGUGUGCGCGCACAUUUAGACUGAGAUAUCUGCCAACAACAACAUAUACCGAUGGCCAUACACACGCACAC-CCCGCGCACA ((.(---(((((((((((((((.(((((.((((......))))...))).........(((......))))))))))))))))))))-).))...... ( -34.30) >consensus CGUGG__GUGUGUGUGUGCGUGUGAGCA_UUUUAGACUGAGAUAUCUGCCAACAACAACAAAUACCGAUGCCCACACACAGGCACACACCCACGCACA .((((.(((((((.((((.(((.(.(((....((((........))))..........(.......).))))))).)))).))))))).))))..... (-20.09 = -21.73 + 1.64)


| Location | 18,713,311 – 18,713,408 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 86.25 |
| Mean single sequence MFE | -41.97 |
| Consensus MFE | -28.88 |
| Energy contribution | -31.11 |
| Covariance contribution | 2.23 |
| Combinations/Pair | 1.06 |
| Mean z-score | -4.59 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.945703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
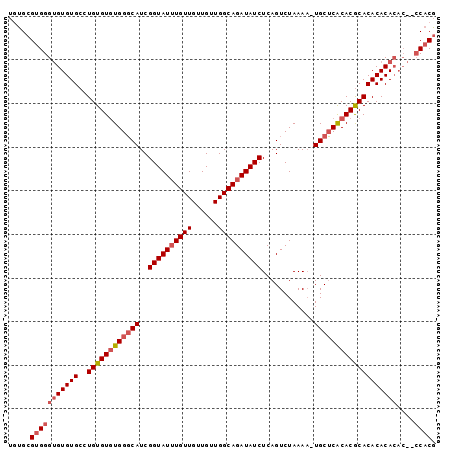
>X_DroMel_CAF1 18713311 97 - 22224390 UAUGCGUGGGUGUGUGCCUGUGUGUGGGCAUCGGUAUUUGUUGUUGUUGGCAGAUAUCUCAGUCUAAAA-UGCUCACACGCACACACACACACCCACG ....((((((((((((..(((((((((((((.((((((((((......))))))))))..........)-))))))))))))....)))))))))))) ( -48.80) >DroSec_CAF1 14709 92 - 1 UGAGCGUGGGUGUGUGCCUGUGUGUGGGCAUCGGUAUUUGUUGUUGUUGGCAGAUAUCUCAGUCUAAAA-UGCUCACACGCACACACA-----CCACU .....((((((((((((....((((((((((.((((((((((......))))))))))..........)-))))))))))))))))).-----)))). ( -41.40) >DroEre_CAF1 13918 94 - 1 UGUGCGCGGG-GUGUGCGUGUGUAUGGCCAUCGGUAUAUGUUGUUGUUGGCAGAUAUCUCAGUCUAAAUGUGCGCGCACACACACACACAC---CACG ((((.(.(..-(((((((((..(((.((((.(..((.....))..).))))((((......))))..)))..))))))))).).).)))).---.... ( -35.70) >consensus UGUGCGUGGGUGUGUGCCUGUGUGUGGGCAUCGGUAUUUGUUGUUGUUGGCAGAUAUCUCAGUCUAAAA_UGCUCACACGCACACACACAC__CCACG .....(((((((((((..((((((((((((..((((((((((......))))))))))............)))))))))))))))))))....)))). (-28.88 = -31.11 + 2.23)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:34 2006