
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,711,340 – 18,711,433 |
| Length | 93 |
| Max. P | 0.909916 |

| Location | 18,711,340 – 18,711,433 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 76.84 |
| Mean single sequence MFE | -24.27 |
| Consensus MFE | -20.02 |
| Energy contribution | -19.80 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909916 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
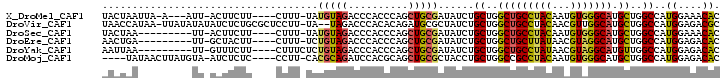
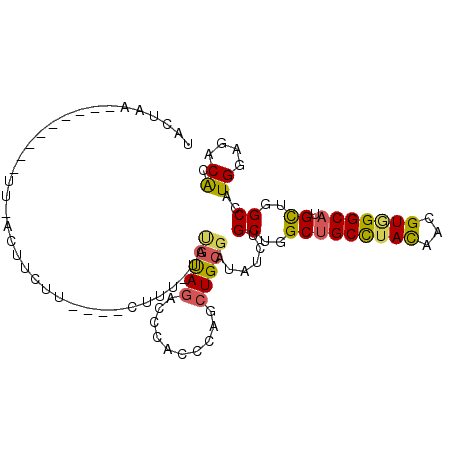
>X_DroMel_CAF1 18711340 93 + 22224390 UACUAAUUA-A---AUU-ACUUCUU----CUUU-UAUGUAGACCCACCCAGCUGCGAUAUCUGCUGGCUGCCUACAAUGUGGGCAUGCUGGCCAUGGAAACAC .........-.---...-.......----....-..(((((..........)))))......((..(((((((((...))))))).))..))..((....)). ( -23.10) >DroVir_CAF1 9534 99 + 1 UAACCAUAA-UUAUAUAUAUCUCUGCGCUCCUU-UA--UAGACCCACACAGAUGCGCUAUCUGCUGGCUGCCUACAACGUUGGCAUGCUGGCCAUGGAGACGC .........-..............(((((((..-..--...........(((((...)))))((..((((((.........)))).))..))...)))).))) ( -25.20) >DroSec_CAF1 12751 88 + 1 UACUAA---------UU-ACUUCUU----CUUU-UAUGUAGACCCACCCAGCUGCGAUAUCUGCUGGCUGCCUACAAUGUGGGCAUGCUGGCCAUGGAAACAC ......---------..-.......----....-..(((((..........)))))......((..(((((((((...))))))).))..))..((....)). ( -23.10) >DroEre_CAF1 11970 88 + 1 AACUGA---------UU-GCUACUU----CUUU-UCUGUAGACCCACCCAGCUGCGAUAUCUGCUGGCUGCUUAUAACGUAGGCAUGCUGGCCAUGGAGACAC ..(((.---------.(-(((((..----....-...))))...))..)))...........((..(((((((((...))))))).))..))..((....)). ( -20.70) >DroYak_CAF1 12205 89 + 1 AAUUAA---------UU-GUUUCUU----CUUUCUCUGUAGACCCACCCAGCUGCGAUAUCUGCUGGCUGCCUAUAACGUAGGCAUGUUGGCCAUGGAGACAC ......---------.(-((((((.----.......(((((..........)))))......((..(((((((((...))))))).))..))...))))))). ( -24.20) >DroMoj_CAF1 9571 93 + 1 ----UAUAACUUAUGUA-AUCUCUC----CCUU-CACGCAGAUCCACGCAGCUGCGCUACCUGCUGGCCGCCUACAAUGUGGGCAUGCUGGCCAUGGAGACAC ----.............-...((((----(...-...((((.....(((....)))....))))(((((((((((...)))))).....))))).)))))... ( -29.30) >consensus UACUAA_________UU_ACUUCUU____CUUU_UAUGUAGACCCACCCAGCUGCGAUAUCUGCUGGCUGCCUACAACGUGGGCAUGCUGGCCAUGGAGACAC ....................................(((((..........)))))......((..(((((((((...))))))).))..))..((....)). (-20.02 = -19.80 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:32 2006