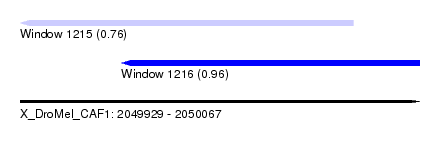
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,049,929 – 2,050,067 |
| Length | 138 |
| Max. P | 0.960219 |

| Location | 2,049,929 – 2,050,044 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 86.75 |
| Mean single sequence MFE | -22.61 |
| Consensus MFE | -20.09 |
| Energy contribution | -19.42 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2049929 115 - 22224390 AUUCAUCCAGUCAGCCCAUCAUUUUUGUAUUUCCCCUGCAAUCGCAAUCGAACUUUGCCCAUGCAGCCUUACUGCAGCUCAAAUUACAAAUCGAUUAGGCUCGCGUGCCAUCCAA .........((.((((.(((...((((((........((....((((.......))))...(((((.....)))))))......))))))..)))..)))).))........... ( -22.84) >DroSec_CAF1 16365 102 - 1 ------------ACCCCGUCACUUUUGCUUUUCCCCUGCAAUCGCAAUCGAACUUUGCCCAUGCAGCC-UACUGCAGCUGAAAUUGCAAAUCGAUUAGGCUCGCGUGCCACCCAA ------------.....(((...(((((.((((....((....((((.......))))...(((((..-..))))))).))))..)))))..)))..(((......)))...... ( -22.70) >DroSim_CAF1 14171 102 - 1 ------------ACCCCAUCAUUUUUGCUUUUCCCCUGCAAUCGCAAUCGAACUUUGCCCAUGCAGCC-UACUGCAGCUGAACUUGCAAAUCGAUUAGGCUCGCGUGCCACCCAA ------------............((((.........))))....((((((..(((((.....((((.-.......)))).....))))))))))).(((......)))...... ( -22.30) >consensus ____________ACCCCAUCAUUUUUGCUUUUCCCCUGCAAUCGCAAUCGAACUUUGCCCAUGCAGCC_UACUGCAGCUGAAAUUGCAAAUCGAUUAGGCUCGCGUGCCACCCAA .................(((...(((((.........((....((((.......))))...(((((.....))))))).......)))))..)))..(((......)))...... (-20.09 = -19.42 + -0.66)

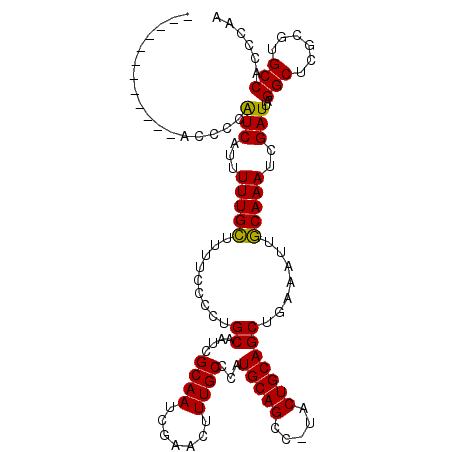
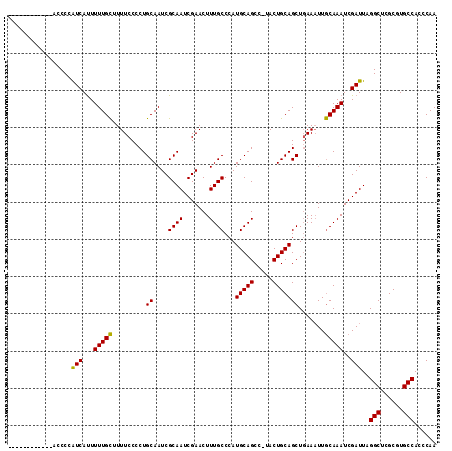
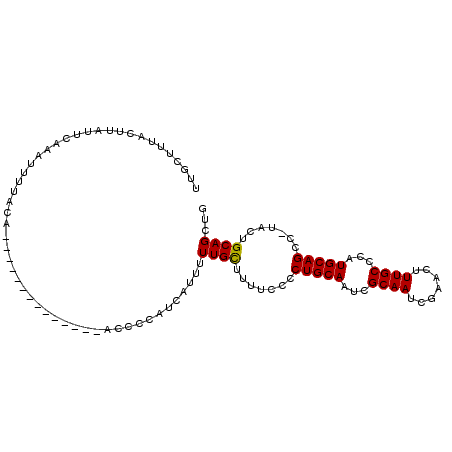
| Location | 2,049,964 – 2,050,067 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 81.05 |
| Mean single sequence MFE | -12.87 |
| Consensus MFE | -12.19 |
| Energy contribution | -11.97 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960219 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2049964 103 - 22224390 U----UUACCUAUUCAAAUUCUGCAAUAUUCAUCCAGUCAGCCCAUCAUUUUUGUAUUUCCCCUGCAAUCGCAAUCGAACUUUGCCCAUGCAGCCUUACUGCAGCUC .----...............(((((.....................((....))........(((((...((((.......))))...)))))......)))))... ( -13.60) >DroSec_CAF1 16400 92 - 1 UUGCUUUACUGAUUCAAAUUUUACA--------------ACCCCGUCACUUUUGCUUUUCCCCUGCAAUCGCAAUCGAACUUUGCCCAUGCAGCC-UACUGCAGCUG ((((......((..((((....((.--------------.....))....))))..))......))))..((((.......))))...(((((..-..))))).... ( -12.70) >DroSim_CAF1 14206 92 - 1 UUGCUUUACUUAUUCAAAUUUUACA--------------ACCCCAUCAUUUUUGCUUUUCCCCUGCAAUCGCAAUCGAACUUUGCCCAUGCAGCC-UACUGCAGCUG .........................--------------............((((.......(((((...((((.......))))...)))))..-....))))... ( -12.32) >consensus UUGCUUUACUUAUUCAAAUUUUACA______________ACCCCAUCAUUUUUGCUUUUCCCCUGCAAUCGCAAUCGAACUUUGCCCAUGCAGCC_UACUGCAGCUG ...................................................((((.......(((((...((((.......))))...))))).......))))... (-12.19 = -11.97 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:54 2006