
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,685,756 – 18,685,852 |
| Length | 96 |
| Max. P | 0.506537 |

| Location | 18,685,756 – 18,685,852 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.76 |
| Mean single sequence MFE | -36.72 |
| Consensus MFE | -17.15 |
| Energy contribution | -17.15 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.47 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
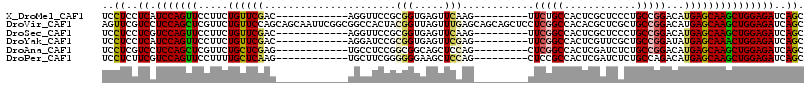
>X_DroMel_CAF1 18685756 96 + 22224390 UCCUCCUCAUCCAGUUCCUUCUGUUCGAC------------AGGUUCCGCGGUGAGUUCAAG---------UUCUGCCACUCGCUCCCUGCCGGACAUGAGCAAGCUGGAGAUCAGC ..((..((.(((((((..(((((((((.(------------(((....((((((.((.....---------....))))).)))..)))).)))))).)))..)))))))))..)). ( -32.90) >DroVir_CAF1 4591 117 + 1 AGUUCGUCCUCCAGCUCGUUCUGUUCCAGCAGCAAUUCGGCGGCCACUACGGUUAGUUUGAGCAGCAGCUCCUCGGCCACACGCUCGCUGCCGGACAUGAGCAAGCUGGAGAUCAGC .....(..((((((((.(((((((.((.(((((....(((.((((((....))......((((....))))...)))).).))...))))).))))).)))).))))))))..)... ( -51.20) >DroSec_CAF1 18048 96 + 1 UCCUCCUCGUCCAGUUCCUUCUGUUCGAC------------AGGUUCCGCGGUGAGUUCAAG---------UUCGGCCACUCGCUCCCUGCCGGACAUGAGCAAGCUGGAGAUCAGC ..((..((.(((((((..(((((((((.(------------(((....((((((.(((....---------...)))))).)))..)))).)))))).)))..)))))))))..)). ( -33.20) >DroYak_CAF1 5157 96 + 1 UCCUCCUCAUCCAGUUCCUUCUGUUCGAC------------AGGAUCCGCGGUGAGUUCGAG---------UUCGGCCACUCGUUCGCUGCCGGAUAUGAGCAAACUGGAGAUCAGC ..((..((.(((((((.....((((((..------------...((((((((((((..((((---------(......)))))))))))).))))).)))))))))))))))..)). ( -33.40) >DroAna_CAF1 7241 96 + 1 UCCUCGUCCUCCAGCUCGUUCUGCUCGAG------------UGCCUCCGGCGGCAGCUCCAG---------CUCGGCCACUCGAUCUCUGCCGGACAUGAGCAAGCUGGAGAUCAGC .....(..((((((((.((((.(((.(((------------((((......)))).))).))---------)(((((............)))))....)))).))))))))..)... ( -38.70) >DroPer_CAF1 4801 96 + 1 UCCUCUUCGUCCAGUUCCUUUUGCUCAAG------------UGCUUCGGGGGGAAGCUCCAG---------CUCCGCCACUCGAUCUCUGCCAGACAUGAGCAAGCUGGAGAUCAGC ..((..((.((((((.....(((((((.(------------(((.((((((((.(((....)---------)).).)).))))).....))...)).)))))))))))))))..)). ( -30.90) >consensus UCCUCCUCAUCCAGUUCCUUCUGUUCGAC____________AGCUUCCGCGGUGAGUUCAAG_________CUCGGCCACUCGCUCCCUGCCGGACAUGAGCAAGCUGGAGAUCAGC ..((..((.(((((((.....((((((......................(((.....)))............(((((............)))))...)))))))))))))))..)). (-17.15 = -17.15 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:22 2006