
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,680,076 – 18,680,275 |
| Length | 199 |
| Max. P | 0.718829 |

| Location | 18,680,076 – 18,680,196 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -53.17 |
| Consensus MFE | -40.78 |
| Energy contribution | -41.57 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579454 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18680076 120 - 22224390 GCUAUGCCUGGUGGAUGCGGCCGCCACUGCCGCUGCCUUGAUCUUGGCCAAGGGCAGAUCCGAUUCGGCAUCUUUGUUCAUUGCCGUGGUCGCAAUGGUUGCUGAGCCACCCGAAGUGGC (((((..(.(((((...((((.(((((((((.(((((........)))..)))))))...((((((((((...........))))).)))))...)))).))))..))))).)..))))) ( -51.60) >DroSim_CAF1 5615 118 - 1 GCUAUGCCUGGUGGAAGCGGCCGCCACAGCCGCUGCCUUGAUCU-GGCCAAG-GCAGAUCCGAUUCGGCAUCUUUGUUUAUUGCCGUGGUCGCAAUGGCUGCUGAGCCACCAGCAGUGGC (((((..(((((((...((((.((((..((..((((((((....-...))))-))))....(((((((((...........))))).))))))..)))).))))..)))))))..))))) ( -57.10) >DroYak_CAF1 5458 120 - 1 GCUGUGCCUGGUGGAAGCGGCCGCCACAGCCGCUGCCUUGAUCUUGGCCAGCGGCAGAUCCGAUUCGGCAUCCUUGUUCAUUCCCGUGGUCGCAAUAGCAGCAGAGCCUCCUGUUGCGGC .....(((........((((((((....(((((((((........)).))))))).((((((...))).))).............))))))))....(((((((......)))))))))) ( -50.80) >consensus GCUAUGCCUGGUGGAAGCGGCCGCCACAGCCGCUGCCUUGAUCUUGGCCAAGGGCAGAUCCGAUUCGGCAUCUUUGUUCAUUGCCGUGGUCGCAAUGGCUGCUGAGCCACCAGAAGUGGC (((((..(.(((((...((((.((((..((..((((((((........))).)))))....(((((((((...........))))).))))))..)))).))))..))))).)..))))) (-40.78 = -41.57 + 0.78)



| Location | 18,680,116 – 18,680,236 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.28 |
| Mean single sequence MFE | -37.83 |
| Consensus MFE | -28.00 |
| Energy contribution | -29.33 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18680116 120 + 22224390 UGAACAAAGAUGCCGAAUCGGAUCUGCCCUUGGCCAAGAUCAAGGCAGCGGCAGUGGCGGCCGCAUCCACCAGGCAUAGCAUGGAAAAGGAGACCAACAUCAGUUCGGGCAGUAGUGCCU .((((....(((((.....((((.((((.((((......))))))))(((((.......)))))))))....))))).(.(((.....(....)...)))).))))(((((....))))) ( -38.50) >DroSim_CAF1 5655 118 + 1 UAAACAAAGAUGCCGAAUCGGAUCUGC-CUUGGCC-AGAUCAAGGCAGCGGCUGUGGCGGCCGCUUCCACCAGGCAUAGCAUGGANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN ....((...(((((......(((((((-....).)-)))))..((.((((((((...)))))))).))....)))))....))..................................... ( -33.30) >DroYak_CAF1 5498 120 + 1 UGAACAAGGAUGCCGAAUCGGAUCUGCCGCUGGCCAAGAUCAAGGCAGCGGCUGUGGCGGCCGCUUCCACCAGGCACAGCAUGGAAAAGGAGACCAACAUCAGUUCGGGCAGUAGCGCCU .((((...(((((((....((.....))(((((((........((.((((((((...)))))))).))....))).)))).)))....(....)...)))).))))((((......)))) ( -41.70) >consensus UGAACAAAGAUGCCGAAUCGGAUCUGCCCUUGGCCAAGAUCAAGGCAGCGGCUGUGGCGGCCGCUUCCACCAGGCAUAGCAUGGAAAAGGAGACCAACAUCAGUUCGGGCAGUAG_GCCU ....((...(((((......(((((((.....))..)))))..((.((((((((...)))))))).))....)))))....)).......................((((......)))) (-28.00 = -29.33 + 1.33)

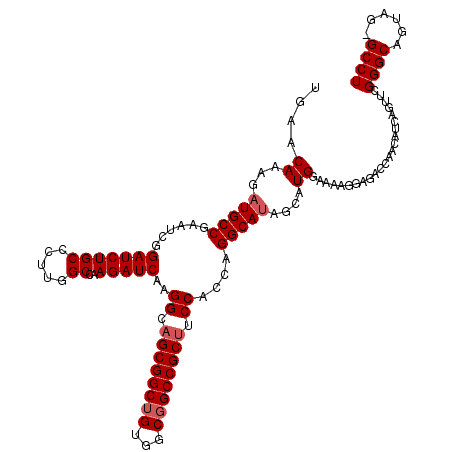
| Location | 18,680,156 – 18,680,275 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 55.99 |
| Mean single sequence MFE | -34.77 |
| Consensus MFE | -21.90 |
| Energy contribution | -23.38 |
| Covariance contribution | 1.47 |
| Combinations/Pair | 1.04 |
| Mean z-score | -0.75 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18680156 119 + 22224390 CAAGGCAGCGGCAGUGGCGGCCGCAUCCACCAGGCAUAGCAUGGAAAAGGAGACCAACAUCAGUUCGGGCAGUAGUGCCUCCGCUUCCAGCAAGGCCAAUUCCGCGGAGAUGCAGCGCA- ....((.(((((.......)))(((((.....(((...((.(((((..((((.(((((....))).))(((....)))))))..)))))))...)))...((....))))))).)))).- ( -41.30) >DroSim_CAF1 5693 120 + 1 CAAGGCAGCGGCUGUGGCGGCCGCUUCCACCAGGCAUAGCAUGGANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN ...((.((((((((...)))))))).)).(((.((...)).)))............................................................................ ( -20.10) >DroYak_CAF1 5538 119 + 1 CAAGGCAGCGGCUGUGGCGGCCGCUUCCACCAGGCACAGCAUGGAAAAGGAGACCAACAUCAGUUCGGGCAGUAGCGCCUCCGCCUCCAGCAAGGCCAAUUCCGCGGAGAUGCAGCGCA- ...((.((((((((...)))))))).))....(((...((.((((...((((.(((((....))).))((....))..))))...))))))...)))......(((.........))).- ( -42.90) >consensus CAAGGCAGCGGCUGUGGCGGCCGCUUCCACCAGGCAUAGCAUGGAAAAGGAGACCAACAUCAGUUCGGGCAGUAG_GCCUCCGC_UCCAGCAAGGCCAAUUCCGCGGAGAUGCAGCGCA_ ...((.((((((((...)))))))).)).(((.((...)).)))...........................((.(((.((((((...................)))))).))).)).... (-21.90 = -23.38 + 1.47)

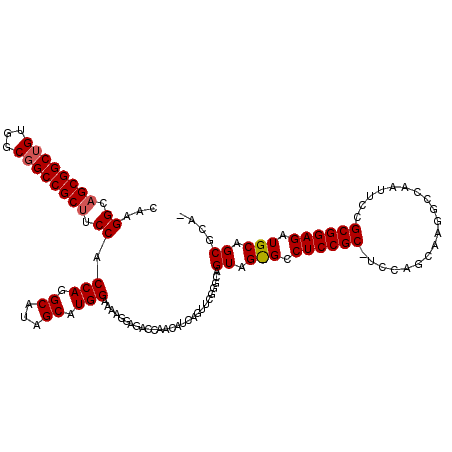

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:20 2006