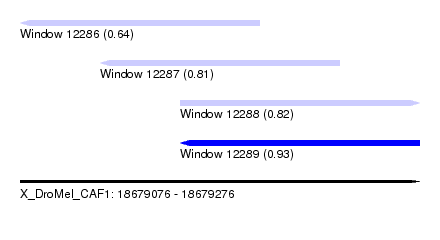
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,679,076 – 18,679,276 |
| Length | 200 |
| Max. P | 0.931547 |

| Location | 18,679,076 – 18,679,196 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -48.77 |
| Consensus MFE | -45.89 |
| Energy contribution | -46.67 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18679076 120 - 22224390 UCCAGAAACUCAAGGGCCGCCUGCACAGCAGGCACAUCUUGCUUGCUCGCCACUUCGCGGGUGAACUCGGUGGCCUCCUUCUUGGGCACCGUAUAGUAGCGGCCAUCCUUGAACGAGCGC .........(((((((((((.(((...(((((((.....)))))))(((((........)))))...(((((.((........)).)))))....))))))))...))))))........ ( -47.20) >DroSim_CAF1 4618 120 - 1 UCCAGAAACUCGAGGGCCGCCUGCACAGCAGGCACAUCUUGCUUGCUCGCCACUUCGCGGGUGAACUCGGUGGCCUCCUUCUUGGGCACCGUGUAGUAGCGGCCAUCCUUGAACGAGCGC .........(((((((((((((((((.(((((((.....)))))))(((((........)))))....((((.((........)).))))))))))..)))))...))))))........ ( -51.20) >DroYak_CAF1 4464 120 - 1 UCCAGAAACUCGAGGGCCGCCUGCACAGCAGGCACAUCCUGCUUGCUCGCCACUUCGCGGGUGAACUCGGUGGCCUCCUUCUUGGGCACCGUGUAAUAGCGUCCGUCCUUGAACGAGCGC .........((((((((.((((((...)))))).......(((..(((((......)))))..)((.(((((.((........)).))))).))....))....))))))))........ ( -47.90) >consensus UCCAGAAACUCGAGGGCCGCCUGCACAGCAGGCACAUCUUGCUUGCUCGCCACUUCGCGGGUGAACUCGGUGGCCUCCUUCUUGGGCACCGUGUAGUAGCGGCCAUCCUUGAACGAGCGC .........(((((((((((((((((.(((((((.....)))))))(((((........)))))....((((.((........)).))))))))))..)))))...))))))........ (-45.89 = -46.67 + 0.78)


| Location | 18,679,116 – 18,679,236 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -48.77 |
| Consensus MFE | -43.62 |
| Energy contribution | -43.73 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.810088 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18679116 120 - 22224390 CAGCGAAUCCCUGUCCCAAUGGGCAGGCAGAACACUGCUGUCCAGAAACUCAAGGGCCGCCUGCACAGCAGGCACAUCUUGCUUGCUCGCCACUUCGCGGGUGAACUCGGUGGCCUCCUU (((((..(((((((((....)))))))..))....))))).............((((((((......(((((((.....)))))))(((((........)))))....)))))))).... ( -46.40) >DroSim_CAF1 4658 120 - 1 CAGAGAAUCCCUGUCCCAAUGGGCAGGCAGAACACUGCUAUCCAGAAACUCGAGGGCCGCCUGCACAGCAGGCACAUCUUGCUUGCUCGCCACUUCGCGGGUGAACUCGGUGGCCUCCUU ..(((.....((((((....))))))((((....))))..........)))(..(((((((......(((((((.....)))))))(((((........)))))....)))))))..).. ( -48.70) >DroYak_CAF1 4504 120 - 1 CAGCGAGUCCCUAUCCCAGUGGGCAGGCAGCACACUGCUGUCCAGAAACUCGAGGGCCGCCUGCACAGCAGGCACAUCCUGCUUGCUCGCCACUUCGCGGGUGAACUCGGUGGCCUCCUU ...(((((.(((((....)))))..((((((.....)))))).....))))).((((((((.(((.((((((.....)))))))))(((((........)))))....)))))))).... ( -51.20) >consensus CAGCGAAUCCCUGUCCCAAUGGGCAGGCAGAACACUGCUGUCCAGAAACUCGAGGGCCGCCUGCACAGCAGGCACAUCUUGCUUGCUCGCCACUUCGCGGGUGAACUCGGUGGCCUCCUU ...................(((((((.(((....)))))))))).........((((((((.(((.((((((.....)))))))))(((((........)))))....)))))))).... (-43.62 = -43.73 + 0.11)


| Location | 18,679,156 – 18,679,276 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -52.23 |
| Consensus MFE | -49.26 |
| Energy contribution | -49.27 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.819701 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18679156 120 + 22224390 AAGAUGUGCCUGCUGUGCAGGCGGCCCUUGAGUUUCUGGACAGCAGUGUUCUGCCUGCCCAUUGGGACAGGGAUUCGCUGUUCGGGCUGACGAACAUCUCGAGCGAUGACGAGGGCGAGA ......(((((((...)))))))(((((((.(((((((((((((((....)).(((((((...))).)))).....))))))))))..)))...((((......)))).))))))).... ( -51.80) >DroSim_CAF1 4698 120 + 1 AAGAUGUGCCUGCUGUGCAGGCGGCCCUCGAGUUUCUGGAUAGCAGUGUUCUGCCUGCCCAUUGGGACAGGGAUUCUCUGUUCGGGCUGACGAACAUCUCGAGCGAUGACGAGGGCGAGA ......(((((((...)))))))((((((((((..(((....((((....))))...(((...))).)))..))))..((((((((.((.....)).)))))))).....)))))).... ( -51.40) >DroYak_CAF1 4544 120 + 1 AGGAUGUGCCUGCUGUGCAGGCGGCCCUCGAGUUUCUGGACAGCAGUGUGCUGCCUGCCCACUGGGAUAGGGACUCGCUGUUCGGCCUCACGAACAUCUCCAGCGAUGACGAGGGCGAGA ......(((((((...)))))))(((((((.(((.(((((((((.....))))(((((((...))).)))).....(.((((((......)))))).)))))).)))..))))))).... ( -53.50) >consensus AAGAUGUGCCUGCUGUGCAGGCGGCCCUCGAGUUUCUGGACAGCAGUGUUCUGCCUGCCCAUUGGGACAGGGAUUCGCUGUUCGGGCUGACGAACAUCUCGAGCGAUGACGAGGGCGAGA ......(((((((...)))))))(((((((.(((.(((((((((((....)).(((((((...))).)))).....)))))))))...)))...((((......)))).))))))).... (-49.26 = -49.27 + 0.01)


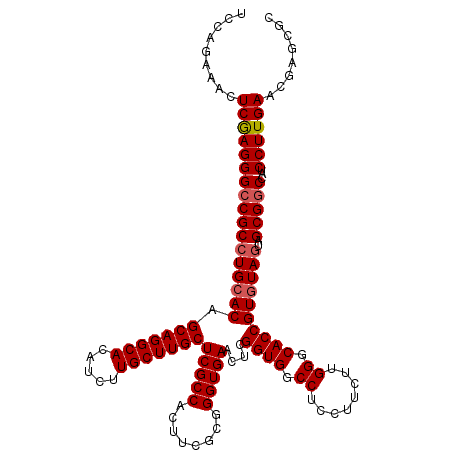
| Location | 18,679,156 – 18,679,276 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -47.59 |
| Consensus MFE | -41.60 |
| Energy contribution | -42.60 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18679156 120 - 22224390 UCUCGCCCUCGUCAUCGCUCGAGAUGUUCGUCAGCCCGAACAGCGAAUCCCUGUCCCAAUGGGCAGGCAGAACACUGCUGUCCAGAAACUCAAGGGCCGCCUGCACAGCAGGCACAUCUU ..((((.((((........)))).((((((......))))))))))..((((.......(((((((.(((....))))))))))........))))..((((((...))))))....... ( -45.46) >DroSim_CAF1 4698 120 - 1 UCUCGCCCUCGUCAUCGCUCGAGAUGUUCGUCAGCCCGAACAGAGAAUCCCUGUCCCAAUGGGCAGGCAGAACACUGCUAUCCAGAAACUCGAGGGCCGCCUGCACAGCAGGCACAUCUU ....(((((((((.((....)))))(((((......))))).(((.....((((((....))))))((((....))))..........))))))))).((((((...))))))....... ( -45.80) >DroYak_CAF1 4544 120 - 1 UCUCGCCCUCGUCAUCGCUGGAGAUGUUCGUGAGGCCGAACAGCGAGUCCCUAUCCCAGUGGGCAGGCAGCACACUGCUGUCCAGAAACUCGAGGGCCGCCUGCACAGCAGGCACAUCCU ....(((((((...((((((....((..(....)..))..))))))((((((.....)).)))).((((((.....))))))........))))))).((((((...))))))....... ( -51.50) >consensus UCUCGCCCUCGUCAUCGCUCGAGAUGUUCGUCAGCCCGAACAGCGAAUCCCUGUCCCAAUGGGCAGGCAGAACACUGCUGUCCAGAAACUCGAGGGCCGCCUGCACAGCAGGCACAUCUU ....(((((((...(((((......(((((......)))))))))).............(((((((.(((....))))))))))......))))))).((((((...))))))....... (-41.60 = -42.60 + 1.00)

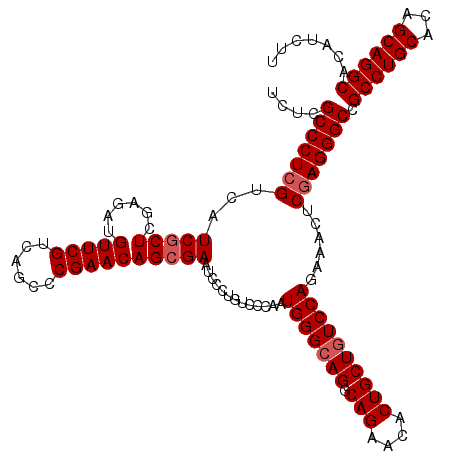
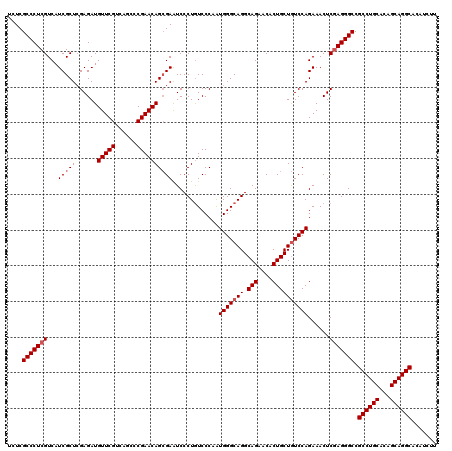
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:15 2006